1. Background
Vaccination is effective in preventing the spread of influenza viruses infections in human and avian populations. Production of egg-based influenza vaccines is still largely implemented in many countries. Despite the considerable advantages of these vaccines, a limited number of mammalian cell lines are approved for more rapid and larger scale cell culture-based vaccine production. As a good alternative substrate, Madin-Darby canine kidney (MDCK) cell line efficiently supports the growth of influenza viruses. However, the cell can be more flexible to supply vaccine production for use in possible influenza pandemic (1, 2). Either in embryonated egg or cell culture, fusion and entry of influenza virus is mediated by specific binding of hemagglutinin (HA) to sialic acid receptors and activation of HA protease (3, 4). The HA0 precursor proteolytically cleaved by intracellular host proteases makes up 2 structurally distinct globular head HA1 and stalk structure HA2 domains (5, 6). Infectivity of influenza viruses depends on forming fusogenic HA molecule and quality of HA0 precursor cleavage. The host proteases responsible for posttranslational cleavage of fusion-incompetent HA0 are expressed in respiratory tract or a limited number of tissue types.
Human and low pathogenic avian viruses have single arginine at HA cleavage site, which is activated by trypsin-like proteases secreted from epithelial cells of the lung or intestinal tissues (7-9). The hydrolysis of HA0 precursor is a necessary step for the viruses to be infective. The ability of the cells to internalize influenza virus is essential for the improvement of viral replication (10, 11). In laboratory trials, adding trypsin or TPCK supports the replication of influenza viruses in a variety of cell cultures. As a protease, the enzyme contributes to viral entry via cleaving HA0, but do not affect the ability of the cells to internalize the virus. However, the viruses grow efficiently on embryonated eggs in the absence of supplemental trypsin (10, 12). It has been found that Factor 10 (FX) is responsible for the cleavage of the fusion proteins of Sendai virus, Newcastle disease virus, and influenza virus (13-15). Considering the high efficiency of FX compared to trypsin on a molar basis and its specificity in cleavage reactions, the genetically manipulated BHK-21/FX cell was established (16). Then, the potential of the cell on virus replication kinetics was evaluated during influenza virus infection process. The results of the data analysis suggested that BHK-21/FX cell provides high-titer replication of low pathogenic influenza virus.
2. Objectives
The present study aimed at applying a technique to establish MDCK/FX manipulated cell, which may provide a new platform for developing influenza vaccine based on the cell culture approaches.
3. Methods
3.1. Transfection of FX Construct onto the MDCK Cell
The cDNA clone of the FX gene was constructed in pcDNA3.1 plasmid (Invitrogen, Carlsbad, CA) according to the previous study (16). In brief, the sequences of chicken FX were deposited in the NCBI GenBank database under accession numbers NM-205022, XM-015277827, and D00844 and were downloaded and aligned. The specific primers were designed based on the open reading frame of FX located in peptidase S1 domain (241-473 nt). MDCK cells were transfected with recombinant vector pcDNA3.1/FX using Lipofectamine 2000 (Life Technologies), and the transfected cells were selected and cultivated in the presence of 10% fetal bovine serum (FBS; Sigma Aldrich, Germany) and 800 mg/mL Geneticin (G418, Invitrogen). After 2 weeks, the survived cells were pooled, passaged 3 times in 1.5 mg of G418/mL, and frozen in aliquots. To verify the expression of FX in MDCK cell, the G418-resistant colonies were subjected to RT-PCR (iNtRON Biotechnology, Korea) using a specific primer set for amplification of FX. The manipulated cell was named MDCK/FX.
3.2. Growth Kinetic Screening of MDCK/FX Cell
The quality of MDCK/FX cell was evaluated by estimating the longevity of cell and the viable cell density in every 17 passages prior to virus inoculation. The cells with concentration of 1 × 106 cells/mL were grown in DMEM (Invitrogen), supplemented with 10% FBS and antibiotic solution, at 37°C with 5% CO2. The rate of culture confluency and the number of live cells were estimated.
3.3. Virus Infectivity Assay
At the 15th passage, the monolayers of MDCK/FX with 80% to 90% confluency were inoculated with H9N2 virus A/chicken/Iran SS8/2011 (17) at MOI of 0.01 PFU/cell. The cultures were incubated up to 72 hours post infection (hpi) at 37°C and controlled for cytopathic effect (CPE) appearance. Four different sets of MDCK culture flasks were inoculated in the presence and absence of supplemental 1 µg/mL TPCK-trypsin (Sigma Aldrich). The trial continued for 7 subsequent passages. The infectivity of influenza virus in each passage was determined using plaque assay and quantitative RT-PCR.
The mRNA level of viral matrix (M) gene from MDCK/FX inoculated cells was analyzed by one-step quantitative RT-PCR (SuperScript TMIII Platinum®Taq Mix, Invitrogen) at a defined interval from 8 hpi up to 48 hpi. The relative expressions were normalized using the housekeeping gene β-actin as internal control. Reactions were repeated 3 times for each sample, and standard deviations (SD) were calculated. The primer sequences (18) used in this study were as follow: MF: 5’-CTCATGGAATGGCTAAAGACA-3’ and MR: 5’-CGATCAADAATCCACAATATC-3’, β-actinF: 5’-TGCTGTGTTCCCATCTATCG-3’ and β-actinR: 5’-TTGGTGACAATACCGTGTTCA-3’.
3.4. Evaluation of Mutations in the Virus
The possible incidence of mutation in viral genome was determined by comparative sequence analysis. The full-length mucleotide sequences of HA genes of influenza virus at 3rd and 7th passages in MDCK/FX were determined and compared to the virus seed (17).
3.5. Statistical Analysis
Data were expressed as mean ± SD. Statistical correlation of data was checked for significance by ANOVA and Student’s t test.
4. Results
4.1. MDCK/FX Cell Establishment and Screening
The plasmid encoded FX was transfected to MDCK cells, and the cells were cultivated under antibiotic selection procedure. After 2 weeks, the survived antibiotic-resistant colonies were selected and subjected to RT-PCR to verify the expression of FX. Specific band at 233 bp lengths has confirmed the presence of FX gene in the cell at each passage.
4.2. Growth Kinetic Characterization of MDCK/FX Cell
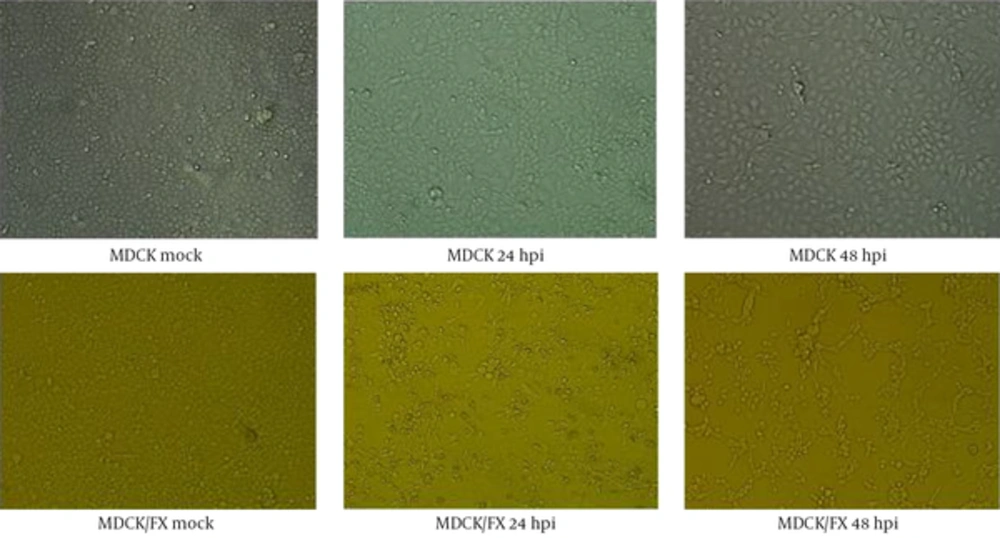
No changes were observed in cell-cell adhesion and cell spreading behavior of MDCK/FX cell following transfection of FX construct. At each passage, the cell grew well and the mean cell density reached to 2 × 105 cells/cm2 after 72 hours of growth. Cell viability remained above 97% during the first day and declined to less than 80% by the 3rd day of incubation, indicating that expression of FX was not toxic to the MDCK cell. The MDCK/FX cell was not different from MDCK cell morphologically.
4.3. Potential Infectivity of Influenza Virus in MDCK/FX
The H9N2 virus replicated well in MDCK/FX and the marked CPEs including giant cell formation were observed at 24 hpi. The massive detaching from the culture flask were found in infected cell at 72 hpi compared to MDCK cell in the presence of supplemental trypsin (Figure 1). No virus growth was found in the absence of trypsin.
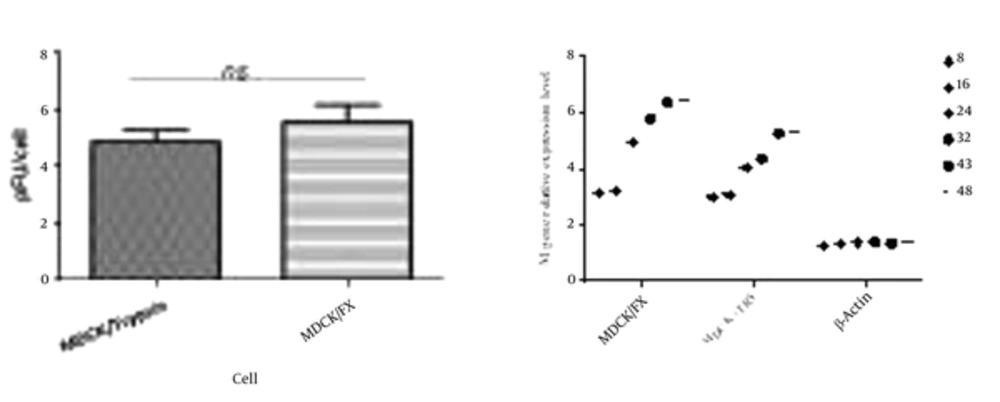
The virus titers peaked higher and earlier in MDCK/FX cell compared to MDCK plus TPCK-trypsin cell; however, no statistical differences were observed (P < 0.01). This clearly indicates that the cell permits the production of infectious virus particles (Figure 2A). The virus showed a modest delay in replication in MDCK cell with supplemental trypsin in which the mean peak titers of the virus in the cell reached after 48 hpi. The replication rate of the virus was monitored by detecting mRNA of M gene in quantitative RT-PCR up to 48 hpi (Figure 2B). In MDCK/FX cell, the numbers of M-expressed cells increased with time elapses due to protease activation of HA cleavage site which supports virus entry and replication. The virus infected-MDCK cells have exhibited the same panel in the presence of trypsin at 8 and 16 hours post infection, but significantly more M gene mRNA copies were found in MDCK/FX cell compared with MDCK cell by 24 hpi (P < 0.01).
4.4. Evaluation of Mutations in the Virus
Nucleotide sequences of HA gene of influenza virus replicated in MDCK/FX were determined at 3rd and 7th passages and the seed virus gene sequence was compared. Mutation in the nucleotide sequences of the viral gene that resulted in the change in amino acid codon was not detected. Sequence analyses of HA glycoprotein showed conservation of residues H183, L190, L226, Q227 and G228 in the receptor binding pocket and RSSR motif in the cleavage site.
5. Discussion
The early detection and isolation of influenza A viruses are very important to design effective control programs and prevent strategy against the infection. Conventionally, the viruses replicate in embryonated eggs and avian or mammalian-origin cells following bind to N-glycan structures of sialic acid (3, 18). It has been suggested that mammalian cell lines vary in the amount of viral particles that attach to the cells. In the present study, we successfully transferred a construct of FX gene to MDCK cell, then, we evaluated the growth kinetics of H9N2 virus on the manipulated MDCK/FX in the absence of TPCK-trypsin at 7 subsequent passages. Virus titration demonstrated that the cell supported high-titer growth of the virus in which the viral titer was comparable to the virus grown in MDCK cells supplemented with TPCK-trypsin. The virus showed a modest delay in replication in MDCK cell. Full-length amplification and sequencing patterns of the HA gene demonstrated that the H9N2 HA gene at 7 passages remained similar to the parent virus. These data indicate the ability of MDCK/FX as a permissive cell to replicate the low pathogenic influenza virus.
It has been reported that some influenza clinical isolates have low growth properties in MDCK cells line (19, 20). Thus, we modeled the embryonated egg cellular factors involved in the viral HA cleavage procedure for use in cell culture. Analysis of the N-glycan in embryonated chicken egg revealed the presence of both α 2, 3 (chorioallantoic) and α 2, 6 (amniotic) linkages, known to be important for efficient virus entry and internalization. During the life cycle of influenza virus, internalization of viral particles into target cells is the primary step for replication and production of high viral titers. Along with the specific receptor distribution of embryonated egg, the FX host cell ubiquitous protease has a role in promoting virus replication. This protease, as a determinant of viral tropisms, cleaves the major glycoprotein and contributes to the fusion activity. Enhancing the membrane fusion leads to increasing the binding rates and virion production.
Catalytic chain (FXa) of the virus activating protease is crucial for the substrate specificity (15), as it cleaves the virus precursor glycoproteins directly and facilitates the virus progeny production and spread (13). The duration of reproductive cycle of influenza virus and release of progeny was estimated to take 8 to 10 hours in MDCK cell and A549 cell (21-23). The progeny virus release time was monitored by estimation of viral M protein expression levels in MDCK/FX cells. Based on the quantitative RT-PCR data, the progeny viruses release in MDCK/FX at 8 hpi was similar to the MDCK cells supplemented with TPCK-trypsin.
Despite similarities of virus replication dynamic, difference in CPE appearance was observed between both cells. CPE appears in a short time in MDCK/FX cell compared to MDCK cells that can result from the effective cleavage of HA protein. The HA cleavage site of low pathogenic influenza virus is activated only in the respiratory and intestinal tract where trypsin-like enzymes are secreted (24). Addition of exogenous trypsin is required for the efficient replication of the viruses in vitro unless serine proteases are expressed in these cells. Analysis of replication kinetics of influenza viruses in different cell lines showed that the viral replication and faster progression of infection in LMH, HepG2, and A549 cells are higher than MDCK, HeLa and Vero cell lines because they are able to activate HA monobasic cleavage site and promote virus entry in the absence of trypsin (1, 11, 21).
It has been suggested that treatment of the cells with trypsin may cause an increase in cell death and a decrease in the virus titer. Moreover, FX is 20 times more effective than trypsin on a molar basis, and 8 times less volume of the factor than trypsin is required for the same trial. Trypsin causes large nonspecific loss of viruses’ infectivity under incubation conditions similar to FX. Thus, the factor is much more effective and specific in cleavage activation than in trypsin (13, 25). The potential of FX to cleave HA and promote replication of influenza viruses is confirmed by our study in which maximum viral titers were reached one day earlier in MDCK/FX with comparative MDCK cells.The impact of FX on H9N2 virus replication kinetics upon engineered expression in BHK-21 cell line has been evaluated (16).
In addition to activation of HA protease processing, type and density of receptors play an important role in virus replication (3, 5). Clinical isolates of human influenza viruses bind strongly to 6-linked sialic acid, whereas avian viruses bind to 3-linkage. The distribution of sialic acid on cell surfaces and affinity of the HA to the receptors are determinants of host tropism. Stable transfection with the SIAT1 gene increased the concentration of the sialyl α2, 6-Gal receptor on the surface of MDCK cells about twofold and decreased the same amounts of α2, 3-Gal than parent MDCK cells (26).
It seems that a low concentration of sialyl α2, 3-Gal receptors in conventional cell lines may lead to non-efficient replication of avian influenza isolates in SIAT-MDCK cell (27). Unlike this cell, amniotic cells of chicken embryonated eggs contain N-glycans with terminal mannose and sulfate residues capable of binding to human and avian influenza viruses. The presence of sulfated glycans may be a reason why human influenza viruses are efficiently replicated in chicken embryonated egg. Once bound, the host FX protease involves in HA priming and proteolytic cleavage. The enzymatically active FXa was detectable only in the allantoic and amniotic fluids (14, 15).
5.1. Conclusions
Using N-glycan structure and FX protease activity of chicken embryonated egg, we introduced a highly permissive engineered cell for influenza virus propagation. The MDCK/FX cell has 2 main features: uniform cell morphology and stable expression of FX. With the establishment of the cell, we will focus on analysis of N-glycans responsible for virus binding, evaluation of susceptibility of the cell to different virus infectious doses, and cultivation of more influenza viruses isolated from human and avian populations in future studies.