1. Background
Neonates have high risk for infection of Klebsiella pneumoniae due to the undeveloped body physiology, resulting in high morbidity and mortality (1, 2). Approximately one million neonatal deaths are caused by infection, which is frequently associated with contaminated medical devices and instrumentation. Factors, like high occupation rates and extensive use of antibiotics, are inevitably leading to the prevalence of drug-resistant strains. As one of the most frequently isolated bacteria in a hospital setting, K. pneumoniae is resistant to many antimicrobial agents by producing a variety of drug-resistance genes, including extended spectrum β-lactamases (ESBLs), AmpC β-lactamases, 16S rRNA methylases, aminoglycoside modifying enzymes and carbapenemases (3).
The increasing number of strains harboring extended-spectrum β-lactamase-producing Enterobacteriaceae (resistant to penicillins, cephalosporins and monobactams) and AmpC β-lactamases is a major treatment challenge. Given the side-effects of antibiotics in newborns, carbapenems are often the only therapeutic option for treating severe infection caused by mutidrug-resistant Enterobacteriaceae (4). In recent years, carbapenem-resistant Enterobacteriaceae in infants and children has become an emerging nosocomial pathogen worldwide.
Carbapenem-resistance is primarily mediated by the production of carbapenemases, which hydrolyze β-lactam of all classes, including the most potent class, carbapenems (5). Molecular structural classifications of carbapenemases include Ambler class A, B and D. The metallo-β-lactamases (MBLs) belong to class B carbapenemases that includes Verona integron-encoded MBL (VIM), IMP (IMP-type MBL) or New Delhi MBL (NDM) (6). Carbapenemases carried on mobile genetic elements, such as transposons or plasmids harboring resistance genes, can transfer into strains capable of efficient human to human transmission (7). The carbapenemase New Delhi metallo-beta-lactamase-1 (NDM-1) is one of the most recently reported MBLs. It was originally identified in India, but now has a global distribution (8).
A few studies on spread of carbapenemase-producing K. pneumonia in newborns have been reported. In August 2011, a nosocomial outbreak of NDM-1-producing K. pneumonia was reported in Colombia involving six patients who were admitted to the neonatal unit of a general hospital in Bogota (9). An outbreak of carbapenemase-producing K. pneumoniae sequence type (ST) 258 was investigated in a neonatal intensive care unit (NICU) in Palermo, Italy and carbapenemase-producing K. pneumoniae was isolated from 10 out of 54 neonates admitted in the outbreak period (10). In China, an outbreak caused by imipenemase-4 (IMP-4)-producing K. pneumonia occurred in a neonatal intensive care unit in 2011 (11). Another outbreak of colonization by carbapenemase-producing K. pneumoniae occurred in a neonatal intensive care unit of Peking union Medical college hospital, China in 2015 (2).
2. Objectives
In this study, our efforts were focused on the 16 isolates from newborns because it poses a more serious problem than adults. The molecular epidemiology of clonally related metallo-β-lactamase-producing K. pneumoniae was investigated by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). The strains were subjected to antimicrobial susceptibility testing and the expression of carbapenemases was investigated to understand the mechanisms responsible for the emergence of carbapenem resistance. Furthermore, plasmid typing was carried out to study the transferability of carbapenem-resistance.
3. Methods
3.1. Ethics Statement
Samples were collected during routine check-ups by medical professionals. The study was approved by the Ethics Committee of Provincial Hospital affiliated with Shandong University and carried out in accordance with the approved guidelines.
3.2. Collection of Isolates
A total of 16 K. pneumonia isolates were recovered from newborns (age of 2 to 28 days) hospitalized in our hospital between September 2011 and June 2014. Hand and nasal swabs were collected from the doctors and nursing staff in the neonatal unit and intensive care unit (ICU), as well as the environmental samples cultured with a swab pre-moistened with sterile saline. The sites for collecting environmental samples included the bed railing, stethoscope, weighing machine, milk powder, suction apparatus, wash basin, radiant warmer, washing area, electric switch, medicine tray and ventilator machine. All isolates were identified on a VITEK-2 compact system (bioMe’rieux, Marcy l’Etoile, France) and screened for carbapenemase production by the modified Hodge test (MTH) and ertapenem-EDTA double-disc synergistic test (12).
3.3. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was determined using the VITEK-2 system (bioMe’rieux, Marcy l’Etoile, France). The susceptibility of meropenem, imipenem, ertapenem, tigecycline, colistin and fosfomycin were verified by the E-test (bioMe’rieux, Marcy l’Etoile, France) method on a Mueller-Hinton agar (Oxoid, Hampshire, United Kingdom). All antibiotics were purchased from Sigma, St Louis, USA. The susceptibility data were interpreted according to the Clinical and Laboratory Standards Institute guidelines, except for colistin and tigecycline, which were interpreted by the 2011 European committee on antimicrobial susceptibility testing break points (available at http://www.eucast.org/clinical_breakpoints/). Escherichia coli ATCC25922 was used for quality control in antimicrobial susceptibility testing.
3.4. Drug-Resistance Genes
Isolates were screened by polymerase chain reaction (PCR) for the presence of the molecular class A carbapenemases (blaKPC, blaGES, blaSME, blaNMC, and blaIMI1) (13-17), class B MBL (blaIMP, blaVIM and blaNDM) (14, 18, 19), class D OXA β-lactamases (blaOXA-48) (15), ESBL genes (blaTEM, blaSHV, and blaCTX-M), plasmid-mediated AmpC b-lactamase genes (blaACC, blaFOX, blaMOX, blaDHA, blaCIT, and blaEBC) and integron (Int1, Int2, Int3) (20-22). All PCR-positive products were sequenced by Shanghai Majorbio Bio-Pharm technology Co. (Shanghai, China), and the results were compared to the sequences available from GenBank (www.ncbi.nlm.nih.gov/blast/).
3.5. Resistance Gene Transfer Experiments
Conjugation transfer assay was performed using azide-resistant Escherichia coli J53 as the recipient strain. Briefly, overnight cultures of the donor strain (100 mL) and recipient strain (200 mL) were mixed with 4 mL fresh Mueller-Hinton broth and incubated at 35°C for 24 hours. The mixture was inoculated on MacConkey agar plates containing sodium azide (100 μg/mL) and imipenem (0.5 μg/mL) at 35°C for 24 hours. Conjugation was confirmed by Indole testing (23). Transconjugants were subjected to PCR to determine the presence of the carbapenemase genes.
3.6. Plasmid Profiling
Bands of high-molecular-weight plasmids were visualized for each isolate by PFGE followed by the S1 nuclease (Takara, Tokyo, Japan) treatment of the genomic DNA embedded in agarose. The DNA fragments were separated on a Chef-DrIII apparatus at 14°C for 20 hours, with a voltage of 6 V/cm, and an initial and final pulse time of 2.16 and 63.8 seconds, respectively (24). The gel was recovered to purify the plasmid bands for further sequencing.
3.7. Genotypes and Homology of Drug-Resistance Isolates
DNA fingerprinting was performed by pulsed-field gel electrophoresis (PFGE). DNA fragments were separated on 1% agarose gels (SeaKem Gold Agarose, Rockland, ME) using a Chef Mapper apparatus (Bio-Rad, Hercules, CA). Electrophoresis was conducted in Tris-boric acid-EDTA (TBE) buffer (0.5 ×) at 14°C and a 6 V/cm gradient with alternating pulses at 120°C and a 2 - 40 seconds pulse time gradient for 22.5 hours. The Salmonella enterica serotype Braenderup H9812 was used as the DNA marker (24). Multi-locus sequence typing (MLST) was performed with seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) according to the protocol described on the K. pneumoniae MLST website
(http://www.pasteur.fr/recherche/ genopole/PF8/mlst/Kpneumoniae.html).
Experimentally determined DNA sequences were uploaded to the MLST database and allelic numbers and sequence types (STs) were obtained.
4. Results
4.1. Molecular Epidemiology of K. pneumoniae
On Sept 2, 2011, the first case of K. pneumonia was identified from a sputum sample in the pediatric intensive care unit (PICU) of our hospital, China. The patient was diagnosed with pneumonia on admission and was admitted to PICU due to heart failure. After symptoms were relieved, the patient was transferred to the neonatal unit. Breathing machines and trachea cannula were used during the treatment period in the PICU. On Dec 30, 2011, the second case of K. pneumonia was identified in the neonatal unit from a blood sample. Eight strains were isolated from the neonatal unit and the PICU since May 30, 2012, which prompted the initiation of active surveillance of all nurses, doctors and environments in the wards. However, negative surveillance results were obtained. The spread of metallo-β-lactamases-producing K. pneumonia (CR-KP) was controlled temporarily by a restricted visitor policy and daily cleaning of all rooms. However, CR-KP strains were isolated from newborns again two months later. Seven patients infected with K. pneumoniae were identified from Oct 2012 to Oct 2013.
In total, 16 isolates were obtained from the neonates in the neonatal unit and PICU in this hospital, during this period, including 9 boys and 7 girls. Table 1 lists the clinical characteristics of these 16 K. pneumonia, including isolation date, specimen and ward distribution. Eighteen patients suffered from pneumonia, two with umbilical infection and one with coronary heart disease. There were 14 premature patients with low birth weight and five were intrauterine infection at birth. During hospitalization, all patients were treated with multiple courses of antimicrobial drugs including cloxacillin, cefathiamidine, ceftizoxime and cefoperazone/tazobactam. Among these patients, ten were treated with carbapenems (imipenem or meropenem).
| Isolate No. | Patient Age (d) | Sex | Ward | Isolate Date | Specimen | PFGE Pattern | ST | EDTA | HTM | Resistance Gene |
|---|---|---|---|---|---|---|---|---|---|---|
| E4 | 10 | M | Neonatal unit | 2011-12-30 | blood | D | 290 | + | - | IMP-8, TEM-1, CTXM-14 |
| E5 | 28 | F | Neonatal unit | 2012-5-17 | sputum | A | 54 | + | - | NDM-1, TEM-1, CTXM-14, DHA-1 |
| E6 | 23 | F | Neonatal unit | 2012-5-18 | sputum | A | 54 | + | - | NDM-1, TEM-1, CTXM-14, DHA-1 |
| E7 | 18 | M | Neonatal unit | 2012-5-24 | sputum | A | 54 | + | - | NDM-1, TEM-1, CTXM-14, DHA-1 |
| E8 | 2 | F | Neonatal unit | 2012-6-5 | sputum | A | 54 | + | - | NDM-1, TEM-1, CTXM-14, DHA-1 |
| E9 | 13 | M | Neonatal unit | 2012-7-27 | sputum | A | 54 | + | - | NDM-1, TEM-1, CTXM-14, DHA-1 |
| E11 | 13 | M | PICU | 2012-8-28 | sputum | C | 705 | + | + | IMP-4, TEM-1, CTXM-14 |
| E12 | 5 | M | PICU | 2012-8-28 | sputum | C | 705 | + | - | IMP-4, TEM-1, CTXM-14 |
| E14 | 14 | F | Neonatal unit | 2012-10-4 | sputum | B | 54 | + | - | IMP-4, TEM-1, CTXM-15, DHA-1 |
| E15 | 1 | F | Neonatal unit | 2012-10-16 | sputum | A | 54 | + | - | IMP-4, TEM-1, CTXM-15, DHA-1 |
| E16 | 3 | M | Neonatal unit | 2012-11-5 | umbilicus | A | 54 | + | - | IMP-4, TEM-1, CTXM-15 |
| E17 | 4 | F | Neonatal unit | 2012-11-9 | sputum | A | 54 | + | - | IMP-4, TEM-1, CTXM-15, DHA-1 |
| E18 | 10 | M | Neonatal unit | 2013-1-30 | sputum | A | 54 | + | - | IMP-4, TEM-1, CTXM-15 |
| E20 | 16 | M | Neonatal unit | 2013-2-27 | umbilicus | A | 54 | + | - | IMP-4, TEM-1, CTXM-15, DHA-1 |
| E21 | 25 | F | PICU | 2013-8-31 | sputum | E | 20 | + | - | NDM-1, TEM-1, CTXM-15, DHA-1 |
| E22 | 17 | M | Neonatal unit | 2013-10-17 | sputum | A | 54 | + | - | IMP-4, TEM-1, CTXM-15, DHA-1 |
Abbreviations: CTXM-14, CTX-M-14 Lactamase; CTXM-15, CTX-M-15 Lactamase; DHA-1, DHA-1 Beta-Lactamase; EDTA, EDTA Synergistic Test; F, Female; , HTM, Modified Hodge Test; IMP-4, Imipenemase-4, IMP-8, Imipenemase-8; M, male; NDM-1, New Delhi Metallo-Beta-Lactamase-1; PICU, Pediatric Intensive Care Unit; ST, Sequence Type; TEM-1, TEM1-Beta-Lactamase.
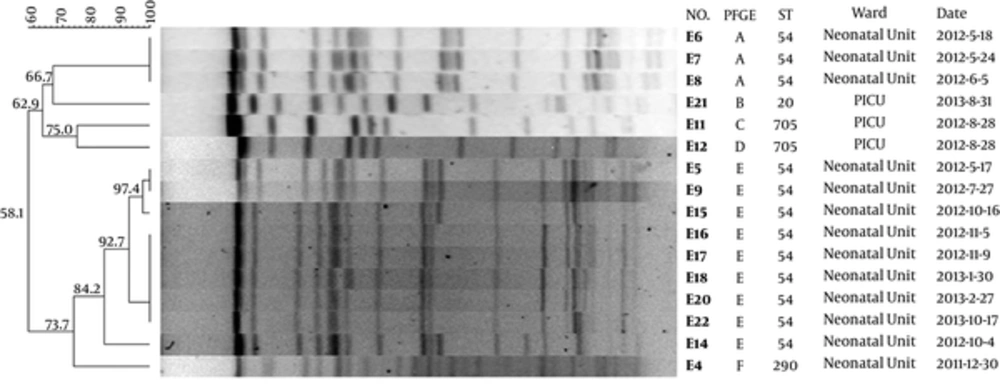
Molecular typing by PFGE (Figure 1) identified 5 major PFGE groups (A to E), among which, type A is the predominate type; 11 isolates from the neonates belonged to type A and all of them were detected in the neonatal unit. The single strain belonging to type B and E was detected in the neonatal unit too. Three strains were isolated from the PICU, two belonged to type C and one to type E.
The dendrogram was generated with the Fingerprinting II Informatix software package (Bio-Rad Laboratories, Hercules, CA) showing fingerprint relationships (XbaI-PFGE) for 16 K. pneumonia strains. The phylogenetic tree was constructed using the Dice coefficient and UPGMA clustering. A genetic similarity index scale is shown in the left of the dendrogram. The PFGE types, sequence type, strain number, collection date and ward distribution are included in each PFGE lane.
Genotyping was carried out by MLST, by amplifying and sequencing seven housekeeping genes. Four different STs were identified, including ST20, ST54, ST290 and ST705 (Figure 1). The MLST results were in good agreement with the PFGE profiles, with the ST54 being the most predominant type. All ST54 isolates were derived from the neonatal unit, out of which, only one strain belonged to PFGE type B and the rest was PFGE type A. ST290 was isolated from the neonatal unit and corresponding to PFGE type D. Two ST705 isolates were from the PICU, which were corresponding to PFGE type C; the other isolate from the PICU were PFGE type E.
4.2. Antimicrobial Susceptibility and Carbapenem Resistance Mechanism
All isolates were resistant to multiple drugs, including cefotaxime, ceftazidime, cefepime, piperacillin-tazobactam and ertapenem. Tigecycline and colistin showed strong activity against K. pneumonia with a susceptibility rate of 100% (Table 2). Three strains that showed low MICs of ertapenem were susceptible to meropenem and imipenem. All isolates from the neonatal unit were resistant to β-lactam antibiotics, but susceptible to trimethoprim-sulfamethoxazole, fosfomycin and amikacin. Strains E11 and E12 from the PICU belonging to ST705 were susceptible to aztreonam, but resistant to fosfomycin, which were the opposite of other strains. All isolates were resistant to ertapenem (MICs > 2 mg/L) and were positive in the EDTA synergistic test. Five of these were positive or weakly positive in the Modified Hodge test (MHT).
| Isolate No. | EPT | IMP | MEM | SXT | TZP | KZ | CTN | CAZ | CRO | FEP | AZM | AK | GN | TOB | CIP | LEV | TGC | CO | FOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E4 | 16 | 3 | 2 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 16 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | 1 | 1 | 0.75 | 0.125 | 64 |
| E5 | > 32 | > 32 | > 32 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 16 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | 1 | 1 | 0.5 | 0.125 | 16 |
| E6 | > 32 | 32 | > 32 | ≤ 1 | 128 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 16 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | 1 | 1 | 0.5 | 0.125 | 32 |
| E7 | > 32 | 16 | > 32 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 16 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | 1 | 1 | 0.75 | 0.125 | 32 |
| E8 | > 32 | > 32 | 32 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 16 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | 0.5 | 0.5 | 0.5 | 0.125 | 16 |
| E9 | > 32 | > 32 | > 32 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 32 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | 1 | 0.5 | 0.75 | 0.125 | 24 |
| E11 | 32 | 8 | 16 | ≥ 16 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 16 | 2 | ≤ 2 | ≥ 16 | 8 | 1 | 1 | 1.5 | 0.125 | 1024 |
| E12 | > 32 | > 32 | > 32 | ≥ 16 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 16 | 2 | ≤ 2 | ≥ 16 | 8 | 1 | 1 | 0.75 | 0.125 | 1024 |
| E14 | > 32 | 4 | 32 | ≤ 1 | 128 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 32 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | 1 | 0.5 | 0.75 | 0.25 | 12 |
| E15 | 4 | 1 | 0.75 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 32 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | 1 | 0.5 | 0.5 | 0.125 | 8 |
| E16 | 32 | 2 | 32 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | ≤ 0.25 | ≤ 0.25 | 0.75 | 0.125 | 12 |
| E17 | 32 | 32 | 32 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | ≤ 0.25 | ≤ 0.25 | 0.5 | 0.125 | 16 |
| E18 | 4 | 0.5 | 1 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 32 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | ≤ 0.25 | ≤ 0.25 | 0.5 | 0.125 | 24 |
| E20 | > 32 | > 32 | 32 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | ≤ 0.25 | ≤ 0.25 | 0.5 | 0.125 | 24 |
| E21 | > 32 | 16 | > 32 | ≤ 1 | 128 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≤ 2 | ≤ 1 | 8 | 1 | 1 | 0.75 | 0.125 | 64 |
| E22 | > 32 | 32 | > 32 | ≤ 1 | 64 | ≥ 64 | ≥ 64 | ≥ 64 | ≥ 64 | 16 | ≥ 64 | ≤ 2 | ≥ 16 | 8 | ≤ 0.25 | ≤ 0.25 | 0.5 | 0.125 | 32 |
Abbreviations: AK, Amikacin; ATM, Aztreonam; CAZ, Ceftazidime; CIP, Ciprofloxacin; CN:, Gentamicin; CO, Colisin; CRO, Ceftriaxone; CTN, Ceftatan; EPT, Ertapenem; FEP, Cefepime; FOS, Fosfomycin; IMP, Imipenem; KZ, Cefazolin; LEV, Levofloxacin; MEM, Meropenem; SXT, Trimethoprim-Sulfamethoxazole; TGC, Tigecycline; TOB, Tobramycin; TZP, Piperacillin-Tazobactam.
Among the carbapenem resistant K. pneumoniae strains, 6 harbored the blaNDM-1 gene and 12 harbored the blaIMP-4 gene. In particular, the E10 strain carried both genes. Other β-lactamases (blaCTX-M-15, blaCTX-M-14, blaDHA-1 and blaTEM-1) were also present in 7 (41.2%), 7 (41.2%), 12 (70.6%) and 17 (100%) strains, respectively. Klebsiella pneumoniae ST705 harbored the same resistant genes including blaIMP-4, blaTEM-1, and blaCTX-M-14 as well as the class 1 gene cassette.
4.3. Conjugative Transfer of Carbapenem Resistance and Plasmid Typing
PFGE analysis revealed that all K. pneumoniae ST54 strains contained three plasmids of ~300 kb, ~200 kb and ~140 kb. The ST705 strain contained two plasmids of ~260 kb and ~55 kb. Conjugation experiments of K. pneumoniae ST54 failed to demonstrate the transferability of carbapenem-resistance from the isolates to the recipient. On the other hand, the K. pneumoniae ST705 transconjugants acquired a single high-molecular-weight plasmid of ~55 kb. Gel recovery was used to purify the three plasmid bands of the K. pneumoniae ST54 strain and the one plasmid band of the K. pneumoniae ST705 transconjugants for further sequencing. The results showed that the blaNDM-1 and blaIMP-4 genes carried by the K. pneumoniae ST54 stains were located on the 300 kb and 200 kb plasmids; the blaIMP-4 gene carried by the K. pneumoniae ST705 strain was located on the 55kb plasmid. All E. coli transconjugants had significantly reduced carbapenem susceptibility and positive EDTA synergistic tests. The recipient carried the same carbapenemase genes as the donor shown by sequencing.
5. Discussion
In this study, we investigated the molecular epidemiology of 16 isolates from newborns in a hospital in Shandong, China. Klebsiella pneumoniae is an opportunistic pathogen posing to great public health threat, especially to newborns. Logan reviewed the clinical history of children with K. pneumoniae infection and noted that the most common condition was pulmonary disease (30%) and premature birth (27%) (7). According to our clinical data, 18 of 21 newborns suffered pulmonary infection. The most common risk factors contributing to newborn carbapenem-resistant K. pneumoniae infection were prematurity, low body weight, intrauterine infection and extensive use of antimicrobial drugs.
There were 14 premature patients with low birth weight, among which, 5 had intrauterine infection at birth. All patients were treated with multiple courses of β-lactam antibiotics including carbapenems. In this study, two patients died and six showed no improvement after treatment. The spread of carbapenem-resistant K. pneumoniae in a hospital is a complex event involving several modes including dissemination of several unrelated strains, the propagation of a single clone from patient to patient and from the environment to patients. There are few reports of outbreaks due to NDM-1-producing Enterobacteriaceae (25-28). Patients with infection and asymptomatic colonization are at risk of invasive infection that often leads to a CRE outbreak in hospitals and communities (29).
In this study, genotyping was carried out by MLST, and the results showed 12 of 13 K. pneumonia isolates from the neonatal unit belonged to the same sequence type, ST54. These isolates displayed the same plasmid profile and antibiotic susceptibility. To understand the transmission mechanisms, we examined the location of the blaIMP-4 and blaNDM-1 genes on the plasmids. We found that the blaIMP-4 gene in the ST705 isolates was transferrable to E. coli recipients through conjugation, while the plasmids in ST54 isolates failed to transfer. The carbapenem-resistant K. pneumoniae strains harbored different types of the blaCTX-M genes; the NDM-1-producing strains carried the blaCTX-M-14 gene, while the IMP-4-producing strains carried the blaCTX-M-15 gene. The ST705 strains were isolated from the PICU, which exhibited a different pattern of antibiotic susceptibility. Specifically, the ST705 were susceptible to aztreonam, but resistant to trimethoprim-sulfamethoxazole and fosfomycin. The sequencing data showed that both ST types harbored the same resistance genes, including blaIMP-4, blaTEM-1, and blaCTX-M-14 as well as the class 1 gene cassette.
Metallo-β-lactamase (MBL)-producing K. pneumoniae is resistant to all ß-lactam agents except aztreonam (30). The ST54 isolates in this study were aztreonam-resistant because they harbored the extended spectrum β-lactamase CTX-M-15. On the other hand, although the ST705 strains also harbored an extended spectrum β-lactamase, these strains were susceptible to aztreonam because the ESBL (CTX-M-14) is less active in hydrolytic activity than CTX-M-15 (31).
5.1. Conclusion
In summary, we investigated the molecular epidemiology of 16 isolates from newborns in a hospital in Shandong, China. The molecular typing was carried out by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). Four ST types were identified, including ST54, ST705, ST20 and ST290. The expression of carbapenemases and the plasmid transferability between ST54 and ST705 were investigated to understand the difference of the antimicrobial susceptibility. To the best of our knowledge, this study is the first report of an outbreak of NDM-1-producing K. pneumoniae in newborns in China. The emergence of the carbapenem-resistant K. pneumoniae isolates underscores the difficulty in eliminating multidrug-resistance bacteria. The outcome of neonatal infections can be improved if the symptoms are identified early and appropriate measures are applied.