1. Background
The increasing antibiotic resistance of Neisseria gonorrhoeae, the pathogen responsible for gonorrhea, is a growing global concern. According to the US Centers for Disease Control and Prevention (CDC), the incidence rate of gonorrhea increased by 111% from 2009 to 2020 (1). Gonorrhea is one of the most common bacterial sexually transmitted infections (STIs), with an estimated 82,400,000 cases worldwide in 2020, as reported by the World Health Organization (WHO) (2). The current empirical treatment for gonorrhea in the United States consists of a combination of ceftriaxone and azithromycin (3). However, the emergence of N. gonorrhoeae isolates with high-level azithromycin resistance (AZM-R) poses a threat to the sustainability of this treatment (4, 5). The rapid emergence of AZM-R has led some countries, such as the United Kingdom, to consider this treatment endangered and recommend ceftriaxone monotherapy instead (3).
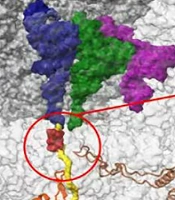
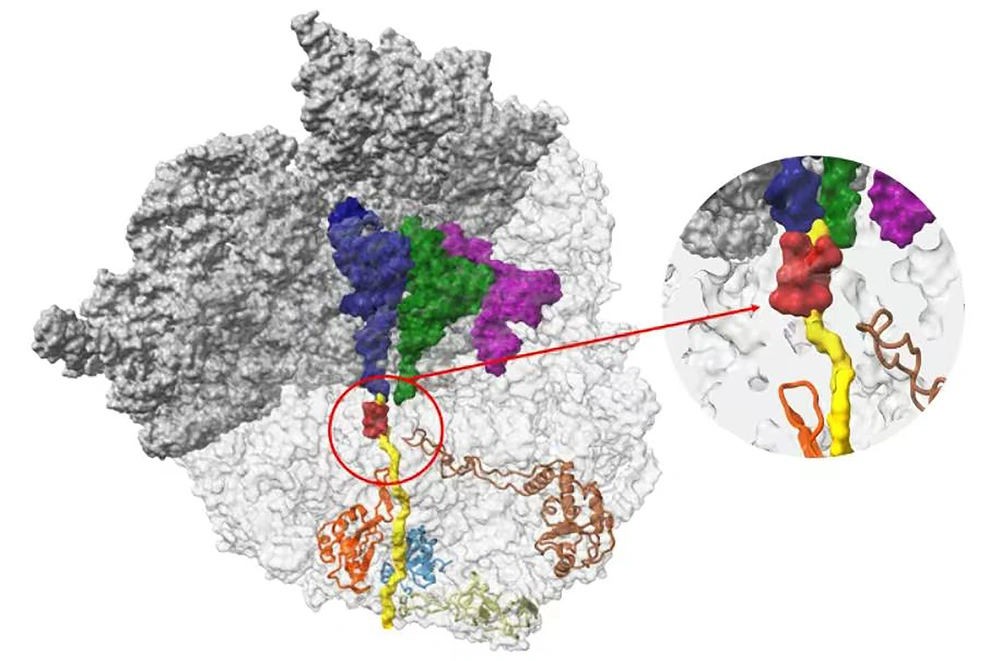
Azithromycin is a 15-membered macrolide that inhibits protein synthesis by binding to the bacterial 50S large ribosome. Azithromycin functions by inhibiting the peptide transfer/translocation step, which blocks the 50S peptide exit tunnel, leading to the release of incomplete peptides from the ribosomes (Figure 1) (6-9). Ribosomal proteins L4 and L22, products of the rplD and rplV genes, respectively, bind to domain I of 23S rRNA to form a channel and facilitate the entry of macrolide antibiotics (10). Mutated L4 and L22 proteins can alter domains II, III, and V, affecting the susceptibility of microorganisms to macrolides (11).
The ribosome nascent chain tunnel environment. Staphylococcus aureus ribosome, where the large subunit is shown in light gray, and the small subunit is shown in dark gray. The A-site, P-site, and E-site docked tRNA molecules are shown in blue, green, and magenta, respectively. The surface of a nascent chain within the peptide exit tunnel is shown in yellow. Bound macrolide is shown in red, and uL4, uL22, uL23, and uL24 are shown in brown, orange, teal, and khaki, respectively. The figure was reproduced with permission from Halfon et al. (9)
Mutations in L4 and L22 have been confirmed to confer resistance to macrolides in various bacteria, such as Brucella, Escherichia coli, and Streptococcus pneumoniae (12-14). Although no mutation related to the 50S ribosome protein L22 has been reported in N. gonorrhoeae, point mutations of the 50S ribosome protein L4, specifically the G70D mutation, have been implicated in macrolide resistance (15, 16). Additional studies have identified other L4 mutations at amino acid positions 68 (G68D and G68C), 69 (T69I), and 70 (G70R, G70S, and G70A), which are associated with decreased susceptibility to azithromycin (16). These mutations are located at the end of the L4 ring near the macrolide binding site and contribute significantly to the development of AZM-R (17).
Multi-locus sequence typing (MLST), a method that characterizes isolates by amplifying sequences from seven housekeeping genes, has been widely used to study the dynamics of N. gonorrhoeae infection and drug-resistant isolates. Multi-locus sequence typing is frequently employed to trace the spread of N. gonorrhoeae isolates, identify lineages, and detect short-term transmission chains (18, 19). It is crucial to deepen our understanding of the emergence and dynamics of drug-resistant N. gonorrhoeae isolates at the national and global levels.
2. Objectives
The objective of this study was to examine the mutation rate and diversity of the rplD gene in N. gonorrhoeae within the Wenzhou area and to assess the extent to which rplD mutations contribute to the consistently elevated azithromycin minimum inhibitory concentration (MIC) levels. Additionally, this study aimed to understand the genetic characteristics of N. gonorrhoeae and analyze the variations in AZM-R among isolates with different genetic backgrounds. Ultimately, the results of this study will serve as a valuable foundation for the prevention and treatment of gonorrhea.
3. Methods
3.1. Bacterial Isolates
This study was conducted within January 2018 and December 2020, during which 37 non-repetitive N. gonorrhoeae isolates were randomly selected from the First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, China. The isolates originated from 5 females and 32 males, aged 18 - 66 years (median age: 31 years), with urine and secretions being the main types of specimens. All experimental isolates were confirmed as N. gonorrhoeae through the fully automated rapid microbial mass spectrometry detection system (VITEK MS, from bioMérieux, France). These isolates included 21 AZM-R isolates and 16 azithromycin-sensitive (AZM-S) isolates.
3.2. Azithromycin Susceptibility Testing
The susceptibility of N. gonorrhoeae to azithromycin was evaluated through MIC determination (Wenzhou Kangtai Biotechnology Co., Ltd., China). All test results were interpreted following the guidelines of the Clinical and Laboratory Standards Institute (CLSI M100ed30) for all antibiotics (20), where MIC ≤ 1 mg/L was deemed sensitive, MIC > 1 mg/L was considered resistant, and MIC ≥ 256 mg/L indicated high-level drug resistance. Individual N. gonorrhoeae isolates were isolated and purified on Thayer-Martin (T-M) selective media. Azithromycin E-test experiments were conducted using GC plates supplemented with 1% IsoVitaleX, vancomycin, colistin, nystatin, and trimethoprim selective supplements in a 5% CO2 incubator at 35°C. This helped determine either the area diameter or MIC. The quality control strain was N. gonorrhoeae ATCC 49226 from the clinical laboratory of the Ministry of Health of the People’s Republic of China. All isolates were stored in glycerol broth at -80°C.
3.3. Extraction of Bacterial Genomic Deoxyribonucleic Acid
The genomic deoxyribonucleic acid (gDNA) of N. gonorrhoeae was extracted through boiling. Moreover, 3 - 5 purified N. gonorrhoeae colonies were suspended in 200 μL of sterile distilled water, boiled in water at 100°C for 10 minutes, and then centrifuged at 13800 g at 4°C for 15 minutes (10). The resulting supernatant contained gDNA, which was stored at -20°C for backup purposes.
3.4. Amplification of the rplD and rplV Genes
The primers for rplD and rplV have been previously published and are listed in Table 1 (10). The polymerase chain reaction (PCR) conditions for rplD involved a pre-denaturation step at 94°C for 4 minutes, followed by denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 45 seconds, with a total of 30 cycles. For rplV amplification, the annealing temperature was adjusted to 55°C, and the extension time was increased to 1 minute; nevertheless, the other conditions remained unchanged (21). The amplified PCR products were subjected to electrophoresis at 110 V for 30 minutes, photographed with a gel imager, and confirmed as single bands. Subsequently, the PCR products were purified using magnetic beads by the sequencing company (Beijing Qingke New Industry Biotechnology Co., Ltd. Hangzhou Branch, China). First-generation sequencing technology was employed for sequencing. The sequencing results were compared to DNA sequences using BLAST to identify genetic mutations.
| Designation | PCR Primer Sequence (5ˊ→3ˊ) | Product Size (bp) | Annealing Temperature (°C) | |
|---|---|---|---|---|
| rplD | F | CAGCGATGTTGTAGTTCGT | 482 | 60 |
| R | ACTCAAGTAATCTTGGCGC | |||
| rplV | F | TCAGCGACAATATGGTTGGT | 468 | 55 |
| R | AGCCCAGTCTTTAGTTACC | |||
The rplD and rplV Gene Primer Sequences and Target Fragment Sizes
3.5. Neisseria gonorrhoeae MLST
Neisseria gonorrhoeae isolates in the present study were genotyped using MLST. Seven housekeeping genes, namely adk, abcZ, aroE, fumC, pgm, gdh, and pdhC of N. gonorrhoeae, were amplified and sequenced. Sequence types (STs) were identified using the online database at the Pasteur Institute MLST typing website for N. gonorrhoeae. The evolutionary relationship between the isolates was analyzed using the platform-independent Java software PHYLOViZ at the single-locus variant (SLV) level with the goeBURST algorithm (22).
3.6. Statistical Analysis
SPSS software (version 22.0) was used to statistically analyze the difference in rplD point mutations between the AZM-S and AZM-R groups of N. gonorrhoeae, with a P-value less than 0.05 indicating significance.
4. Results
4.1. Antimicrobial Susceptibility
Among the 37 isolates of N. gonorrhoeae, the MIC values of azithromycin ranged from 0.125 to 256 μg/mL. Sixteen isolates were classified as sensitive, accounting for 43.24% of the total; however, 21 isolates were resistant, making up 56.76%. Two isolates exhibited extremely high MIC values exceeding 256 μg/mL, categorizing them as high-level AZM-R N. gonorrhoeae.
4.2. Comparison of rplD and rplV Genes
The current study analyzed the DNA of N. gonorrhoeae through PCR amplification and identified 10 isolates with rplD point mutations, resulting in a mutation rate of 27.03%. These mutations were found at four specific sites: G70D, G70S, G68D, and A43T (Table 2). It is worth noting that the A43T mutation was detected for the first time in this study. Except for A43T, the remaining mutations were confirmed to be non-synonymous and were located at the end of the L4 loop, which is near the macrolide-binding site. All these mutations were associated with reduced susceptibility to azithromycin, as suggested by previous studies (5, 14, 16).
| Groups | Strain No. | AZM MIC (µg/mL) | rplD Mutation (L4 Protein) | rplV Mutation (L22 Protein) | MLST ST |
|---|---|---|---|---|---|
| AZM-R (n = 21) | 2 | 4 | G70D | No mutation | 9899 |
| 3 | 8 | No mutation | No mutation | 8123 | |
| 5 | 2 | No mutation | No mutation | 7363 | |
| 6 | 4 | No mutation | No mutation | 1901 | |
| 9 | 1.25 | No mutation | No mutation | 1928 | |
| 13 | 16 | G70S | No mutation | 7367 | |
| 16 | 16 | G70S | No mutation | 7367 | |
| 21 | 1.25 | No mutation | No mutation | 7367 | |
| 25 | > 256 | No mutation | No mutation | 1901 | |
| 27 | 2 | G70S | No mutation | 7367 | |
| 29 | 4 | G70D | No mutation | 14421 | |
| 31 | 1.25 | No mutation | No mutation | 7822 | |
| 32 | 2 | G68D | No mutation | 7822 | |
| 34 | 1.25 | No mutation | No mutation | 1901 | |
| 37 | > 256 | A43T | No mutation | 1588 | |
| 38 | 2 | No mutation | No mutation | 1928 | |
| 39 | 1.25 | No mutation | No mutation | 14283 | |
| 41 | 4 | G70D | No mutation | 15251 | |
| 42 | 1.25 | No mutation | No mutation | 8123 | |
| 43 | 2 | G68D | No mutation | 7822 | |
| 44 | 8 | No mutation | No mutation | 8123 | |
| AZM-S (n = 16) | 1 | 0.25 | No mutation | No mutation | 7822 |
| 8 | 1 | No mutation | No mutation | 8123 | |
| 12 | 0.5 | No mutation | No mutation | 7822 | |
| 17 | 0.125 | No mutation | No mutation | 1928 | |
| 18 | 1 | No mutation | No mutation | 7363 | |
| 19 | 0.5 | No mutation | No mutation | 1928 | |
| 20 | 1 | No mutation | No mutation | 1901 | |
| 22 | 0.125 | No mutation | No mutation | 1588 | |
| 23 | 0.032 | No mutation | No mutation | 1600 | |
| 24 | 0.125 | No mutation | No mutation | 1588 | |
| 26 | 0.5 | No mutation | No mutation | 8123 | |
| 28 | 1 | No mutation | No mutation | 1928 | |
| 30 | 0.125 | No mutation | No mutation | 8123 | |
| 33 | 0.25 | G70S | No mutation | 7367 | |
| 36 | 0.25 | No mutation | No mutation | 1600 | |
| 40 | 0.5 | No mutation | No mutation | 8123 | |
| P | 0.015 |
Characterization and Molecular Typing Results of Clinical Isolates of Neisseria gonorrhoeae
When comparing the AZM-R and AZM-S groups based on whether the MIC value of azithromycin exceeded 1 μg/mL, it was observed that the point mutation rate of rplD in the AZM-S group was significantly lower than in the AZM-R group (P < 0.05). This indicates that the presence of point mutations in rplD was more common in isolates resistant to azithromycin than those sensitive to azithromycin. Another noteworthy discovery was that one of the two high-level AZM-R isolates exhibited an rplD A43T mutation; nonetheless, no mutations were detected in the other. Lastly, the study did not identify any rplV mutations in the analyzed isolates.
4.3. Molecular Epidemiologic Typing
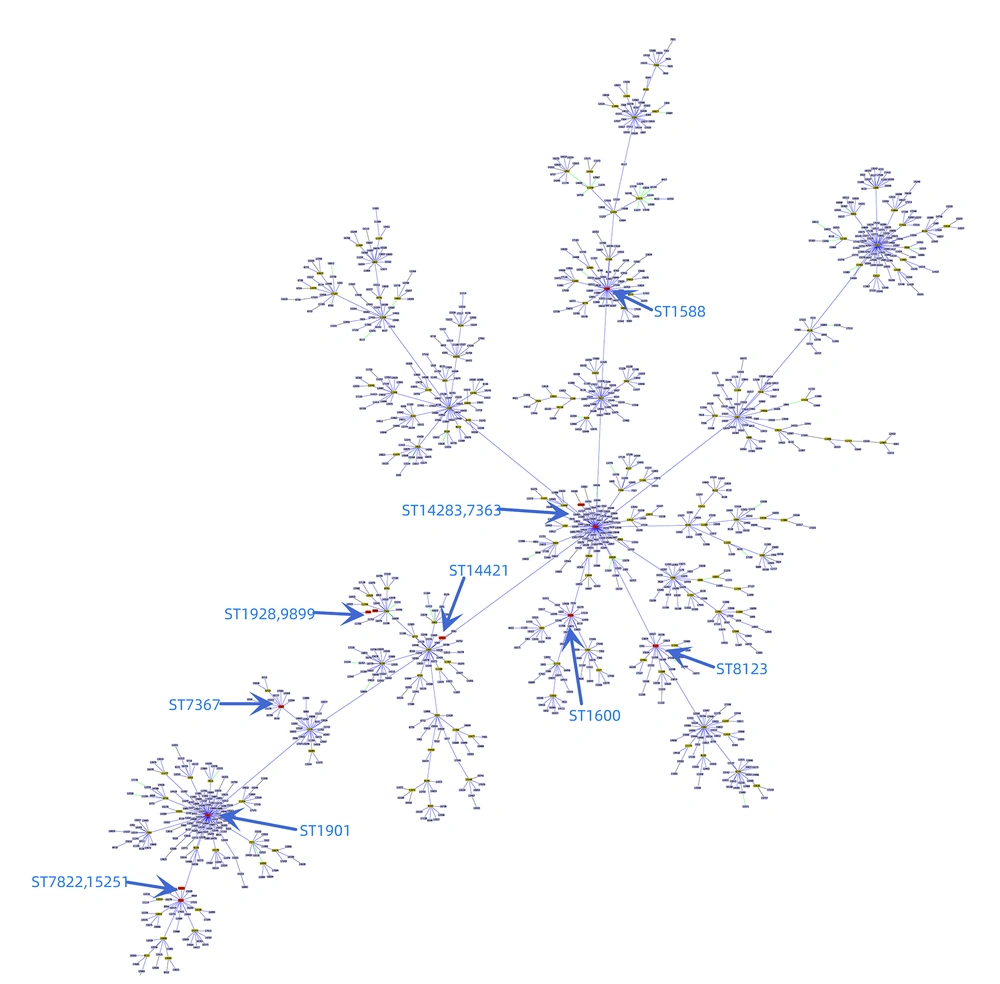
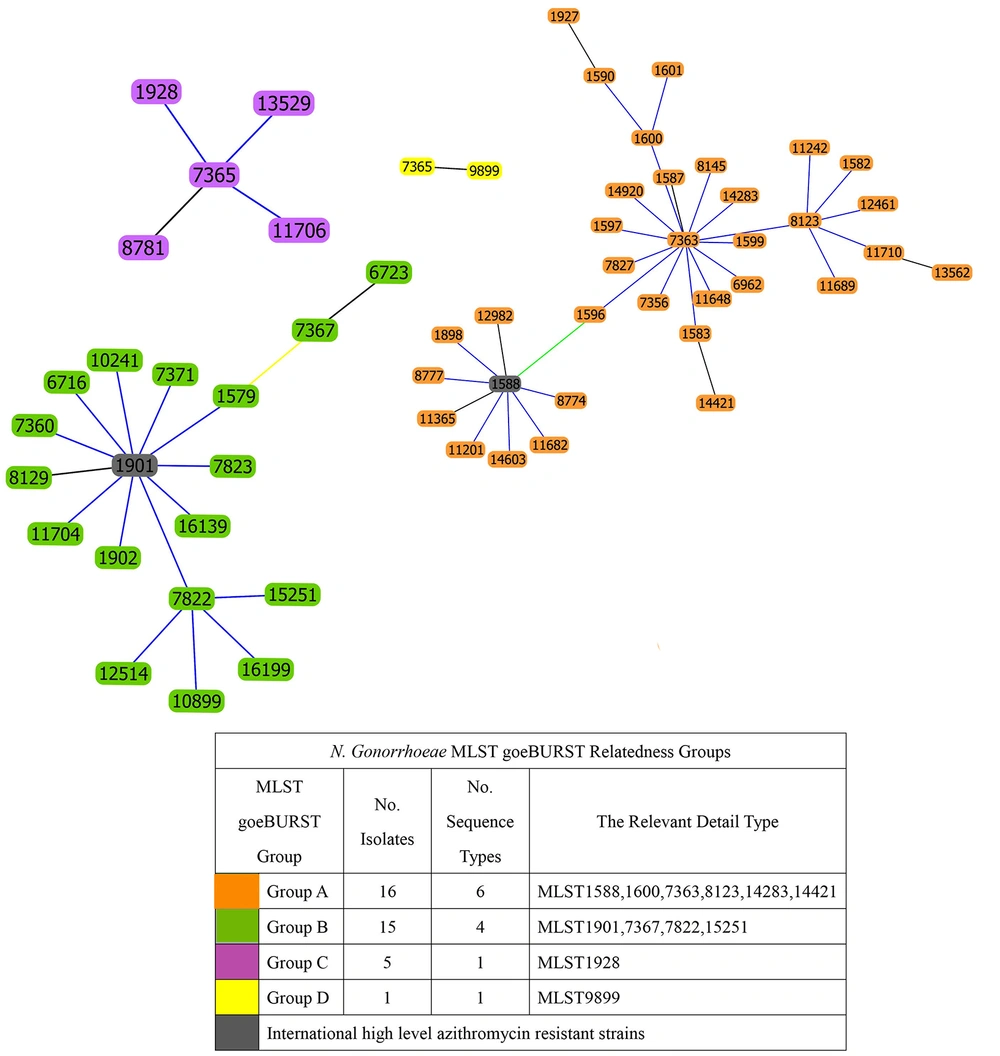
Among the 37 clinical isolates of N. gonorrhoeae, MLST identified 12 different STs. The distribution of these STs was as follows: ST8123 accounted for 18.91% (7/37), ST7822 for 13.51% (5/37), ST7367 for 13.51% (5/37), and ST1928 for 13.51% (5/37). The STs of two high-level AZM-R isolates were identified as ST1901 and ST1588. Using goeBURST minimum spanning tree analysis on the MLST allelic spectrum of the seven MLST housekeeping genes of N. gonorrhoeae, the genetic relatedness of the MLSTs in this experiment was compared to all the MLSTs of N. gonorrhoeae (Figure 2). Based on these genetic relationships, the MLSTs identified in this study were categorized into four large clusters, as shown in Figure 3. Cluster A isolates consisted of 6 different ST types centered around ST7363, with ST14283, ST1600, and ST1823 representing single locus variants of ST7363. Cluster B isolates comprised four different ST types centered around ST1901, with ST7822 being a single locus variant of ST1901. Cluster C and D isolates each contained only one type.
The two isolates highly resistant to azithromycin, ST1588 and ST1901, differed at the four loci adk, aroE, fumC, and gdh. They belonged to cluster A and cluster B, respectively, indicating that they originated from distinct genetic backgrounds, as shown in Figure 2. This suggests that high-level AZM-R N. gonorrhoeae can emerge from different genetic backgrounds, potentially necessitating different treatment strategies.
5. Discussion
Over the years, gonorrhea has developed resistance to all classes of antimicrobials targeting it, especially with the emergence of isolates with reduced susceptibility to azithromycin and ceftriaxone, posing a public health concern due to the rise of incurable gonorrhea infections (23). To prevent further drug resistance, a combination of azithromycin and ceftriaxone has been used for the treatment of gonorrhea (23, 24). However, despite combination therapy, azithromycin resistance has continued to increase, and the emergence of N. gonorrhoeae resistant to both azithromycin and ceftriaxone since 2018 is of even greater concern (25). In 2001, N. gonorrhoeae with high-level resistance to azithromycin (MIC ≥ 256 mg/L) was first isolated in Argentina, and it has been constantly reported in various countries, including England, Italy, the United States, Canada, and China (26). The G70D mutation was the most frequently found point mutation in rplD, which is significantly related to increased AZM-R (16). A recent bacterial genome-wide association study confirmed the role of the rplD G70D mutation in mediating macrolide resistance, showing an average six-fold increase in clarithromycin MICs, with the MICs of azithromycin and erythromycin being 4 times higher than that of the wild-type isolates with the same gene (16).
In addition to detecting the rplD G70D mutation, G70S, G68D, and A43T mutations were also detected in this study. Out of the isolates with rplD mutations, only one N. gonorrhoeae strain with the G70S mutation was sensitive to azithromycin; nevertheless, the other isolates with the G70D, G70S, or G68D mutation were resistant to azithromycin. When comparing the AZM-S group to the AZM-R group, it was observed that the point mutation rate of rplD in the AZM-S group was significantly lower than in the AZM-R group (P < 0.05). This was consistent with previous research results. Although the rplD mutation did not significantly increase the MIC value, it played a role in mediating treatment failure caused by the increase of drug resistance sub-breakpoint (27). Various mutations, such as G68D, G68C, T69I, G70D, G70S, G70A, and G70R, reduced the sensitivity of macrolides by narrowing the peptide exit tunnel and interrupting the binding of macrolide antibiotics to the 50S subunit of ribosome (Figure 1) (16, 28, 29). Therefore, the detection and monitoring of these mutations are crucial in the fight against antibiotic-resistant N. gonorrhoeae.
It was generally believed that mutations in the rplD gene were associated with low-level resistance to macrolides (MIC: 0.5 mg/L); nonetheless, mutations in the rplV gene were associated with low to high-level resistance (MIC: 1.5 to 256 mg/L) (5). No mutations of the rplV gene were observed in this study, which is similar to previous research, possibly due to the relatively few N. gonorrhoeae isolates highly resistant to azithromycin. Of particular interest is the detection of the A43T mutation in the rplD gene in a high-level AZM-R N. gonorrhoeae strain, which, to the best of our knowledge, has not been reported before. The presence of this previously undescribed mutation suggests a potential mechanism for the high resistance of N. gonorrhoeae to azithromycin, although further verification is needed. Multiple previously undescribed mutations in rplD were observed to be associated with higher azithromycin MICs than the G70D mutation (16).
Laumen et al. demonstrated the G70D mutation in one strain with a MIC value of 0.5 mg/L, while the R71C mutation in 2 isolates with MIC values of 24 and 32 mg/L, respectively (5). It is suggested that mutations in rplD/rplV serve as stepping stones to higher-level resistance, with mutations first occurring in the ribosomal genes followed by the MtrCDE efflux pump and 23S rRNA (5). Although AZM-R in N. gonorrhoeae is mainly attributed to 23S rRNA point mutations and overexpression of the MtrCDE efflux pump, the A43T mutation found in this study might represent another mechanism for high resistance. Further research is needed to confirm the potential role of this mutation in antibiotic resistance (8, 30, 31). In this study, MLST typing was used to understand the spread of gonorrhea.
Among all identified MLST types, up to 33.33% (4/12) of STs were represented by only a single isolate, indicating that N. gonorrhoeae, clinically isolated in Wenzhou, China, showed considerable genetic diversity. Shimuta et al. reported that MLST ST7363 and MLST ST1901 N. gonorrhoeae showed their capacity to develop high-level in vitro resistance to ceftriaxone and called them “superbugs” (32). In this study, 5 isolates of ST7363 and 4 isolates of ST1901 were sequenced, accounting for 24.32% (9/37), indicating that the proportion of “superbug” in the Wenzhou area was quite considerable, and there was a risk of multidrug-resistant (MDR) N. gonorrhoeae, which needed to be paid great attention to.
The most common MLST type of AZM-R N. gonorrhoeae was observed to be ST1901, consistent with previous reports from Taiwan and China (16, 32). This type, along with ST7365 and ST1927, was observed to be prevalent in Taiwan, with higher MIC values for ceftriaxone and azithromycin reported for ST1901 isolates (16). The 2 isolates of highly AZM-R N. gonorrhoeae isolated in this experiment were ST1901 and ST1588, belonging to cluster B and cluster A in the goeBURST minimum spanning tree, respectively. These types differed at four loci (adk, aroE, fumC, and gdh), indicating significant genomic differences, which are similar to the previous report (10, 26).
Although ST1588 has not been previously reported as a highly AZM-R strain, it was closely related to the “superbug” ST7363, belonging to the same cluster A and differing by only 2 loci (fumC and gdh). Therefore, increased surveillance of this type is also recommended. The international emergence and spread of antibiotic-resistant N. gonorrhoeae emphasize the need to develop epidemiologically relevant surveillance systems, which not only closely track the spread of known resistant isolates but also rapidly detect novel resistance mechanisms. Furthermore, it should develop rapid and reliable genetic methods to predict antibiotic resistance. In a clinical setting, it is of paramount importance to apply appropriate therapies to limit clonal expansion, thereby having the opportunity to counteract the evolution and spread of gonorrhea.
5.1. Conclusions
Although this study has limitations, such as a relatively small overall number of N. gonorrhoeae isolates and a very small number of AZM-R isolates, especially highly resistant isolates, the mutations discovered in the rplD gene, particularly the newly identified A43T mutation, hold great significance in the research on N. gonorrhoeae resistance to azithromycin in East China. It is necessary to be vigilant about the spread of MLST ST1901, ST7363, and ST1588, especially ST1901 clones.