1. Background
Early immune responses to COVID-19 can help eliminate the virus; therefore, strategies to improve the immune system have become important in disease prevention. As the disease progresses, inflammation and fibrosis of the lungs occur due to cytokine release, which worsens symptoms. Vitamin D has been shown to reduce the risk of respiratory tract infections caused by SARS-CoV-2, and COVID-19 patients have been found to have lower levels of plasma 25(OH)D compared to other patients (1-3). Vitamin D plays a crucial role in the innate immune response to SARS-CoV-2 by increasing the expression of the vitamin D receptor, which enables Toll-like receptors to recognize viral dsRNAs; it also induces secretion of antiviral peptides such as cathelicidin and beta defensins-2 that can block viral entry into cells and suppress viral replication. Vitamin D also modulates adaptive immunity. It activates regulatory T cells that secrete IL-10, reducing the anti-infectious response and causing suppression of Th1 and Th17 cells; dominant Th2 cells inhibit the inflammatory process, Th1-mediated cytokines, and TNF. Severe COVID-19 infection is associated with increased production of proinflammatory cytokines and increased risk of pneumonia and acute respiratory distress syndrome (ARDS). However, adequate vitamin D levels can activate Treg cells and contribute to reducing the severity of the disease and the cytokine storm (4, 5).
The genomic effects of vitamin D are mediated by the nuclear vitamin D receptor (VDR). Vitamin D receptor acts as a ligand-dependent transcription factor that modifies the expression of vitamin D-responsive genes (6). Vitamin D receptor is an intracellular steroid hormone receptor that binds to 1,25(OH)2D and interacts with the vitamin D receptor response elements of target genes to produce various biological effects (7). The binding of 1.25 OH D to the VDR translocates the complex into the cell nucleus and regulates the expression of hundreds of genes, including cytokine production (6). Vitamin D can reduce IFN gamma and TNF alpha. In addition, it decreases the release of proinflammatory cytokines secreted by macrophages and enhances the release of IL-10, leading to increased production of anti-inflammatory cytokines (8, 9). The VDR gene is situated on the long arm of chromosome 12 (12q12-14) and contains almost 200 single nucleotide variations (7). The VDR gene has four major polymorphisms (Bsml, Taq1, Apal, and Fok1) that affect VDR mRNA stability, expression, and activity. Notably, Fok1 and Taq1 are the most widely investigated VDR polymorphisms (10).
2. Objectives
The most important role of vitamin D is to shift responses from T helper 1 to T helper 2 and to modulate the immune response and secretion of inflammatory cytokines. In this regard, it's rational to assume that any disruption in the delivery, activation, and function of Vitamin D can directly and indirectly influence the efficient function of Vitamin D in the host immune system. The primary objective of this study was to assess the prevalence of Taq1 and Fok1 polymorphisms in VDR among SARS-CoV-2-infected patients and determine whether they are linked to COVID-19 susceptibility. The secondary objective was to examine the connection between serum vitamin D levels and COVID-19 susceptibility. Furthermore, we intended to investigate whether comorbidities increase the risk of contracting COVID-19.
3. Methods
This study was carried out by Prof. Dr. Cemil Tascioglu City Hospital. The study included 200 participants in total, aged between 18 - 80 years. The patient group in the study comprised 100 COVID-19-positive hospitalized individuals who did not require intensive care, as confirmed by positive PCR and thoracic CT findings. The control group included 100 preoperative patients who tested negative for COVID-19 PCR, had no symptoms compatible with COVID-19, and were undergoing elective surgery. Blood samples were collected and analyzed for Fok1 and Taq1 polymorphisms in the VDR gene and serum 25 (OH)D levels. Patients were identified and recorded based on epidemiological data, including age, gender, comorbidities, drug use (as exclusion criteria), and thorax CT results. Both groups provided written consent and obtained ethics committee approval (Demiroglu Bilim University Clinical Research Ethics Committee [04.08.2020/2020-14-02]) in accordance with the Declaration of Helsinki.
Blood samples were obtained within 48 hours of hospital admission with informed consent and consent form signing and stored appropriately. The study excluded patients with chronic diseases that may affect vitamin D levels, such as obesity, chronic kidney or lung failure, malignancy, autoimmune disease, hyperparathyroidism, hypoparathyroidism, and pituitary endocrine disorders. Individuals who received vitamin D treatment, calcium, multivitamins, or steroid treatment within the last 6 months and those on antiepileptic drugs that might affect vitamin D metabolism were also excluded from the study.
3.1. Detection of Fok1 and Taq1 Polymorphisms
To detect polymorphism, 2 ml peripheral blood samples were collected from both study groups into tubes containing ethylene diamine tetra acetic acid (EDTA). The samples were stored at +4°C until the polymorphism study was conducted. DNA extraction using commercially available kits (Krosgen, Biotechnology) was performed on collected blood samples to perform polymorphism analysis. The DNA concentration was then determined using a Nano-Drop spectrophotometer (Thermo-Scientific, USA), and the extracted DNA samples were stored at -20°C until the polymerase chain reaction (PCR) was conducted.
To perform PCR amplification of Fok1 (rs2228570) and Taq1 (rs731236) polymorphic regions in the VDR gene, separate primers (Primer sequences for FokI rs2228570 were: 5'-ACCGTGGCCTGCTTGCT-3' (forward) and 5'-AGGGTCAGGCAGGGAAGTG-3' (reverse); for TaqI rs731236: Forward primer 5'CAACCAAGACTACAAGTACCGCGTCAGTGA3', Reverse Primer: 5'CAC TTCGAGCACAAGGGGCGTTAGC3') and probes were designed. The gene sequence was obtained from the Ensembl database (https://www.ensembl.org/index.html), and Primer 3 software was used to design the primers. The designed primer sequences were verified using the BLAST software (https://www.ncbi.nlm.nih.gov). The SensiFAST™ Lo-ROX Genotyping Kit (Bioline) was used to perform PCR reactions in a 20 µL final volume containing 10 µL Master Mix, 0.9 µM forward primer, 0.9 µM reverse primer, 0.2 µM for each probe, 1 µL dH20, and 5 µL DNA. Polymorphism was evaluated by analyzing heterozygous and homozygous (mutant) genotypes (11).
3.2. Serum 25 (OH)D Levels Measurements
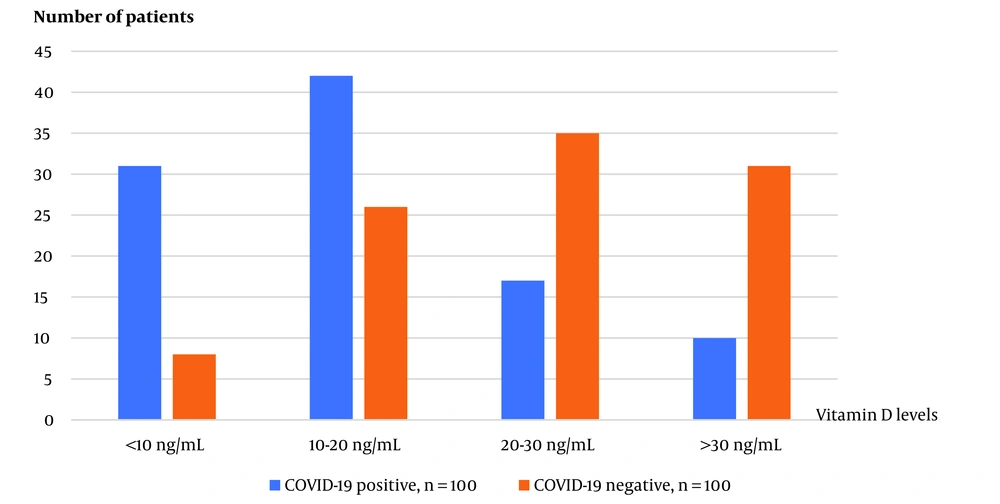
To determine serum 25 (OH)D levels in both study groups, 10 ml blood samples were collected in plastic gel tubes with red caps and stored at -20°C until analysis. The Alinity 25-OH Vitamin D reagent commercial kit (USA) and Abbott (USA) device were used for analysis using the chemiluminescent microparticle immunoassay (CMA) method, following the manufacturer’s instructions. Vitamin D status was defined based on serum levels of 25(OH)D. Levels of at least 30 ng/mL were considered sufficient, 21 - 29 ng/mL were considered insufficient, and levels below 20 ng/mL were classified as deficient. Severe deficiency was defined as levels below 10 ng/mL (12).
3.3. Statistical Analysis
Descriptive analyses were presented using means ± standard deviation for the normally distributed variables. The student's t-test was used for parametric variables like age and level of vitamin D. The Mann-Whitney U test was used for non-parametric variables and was presented with median values of 0.25 and 0.75 percentiles. Chi-square tests are used to compare categorical variables like gender, type 2 DM, hypertension, more than one comorbidity, Fok1, and Taq1 polymorphisms—odds ratio used for risk analyses between two groups. The categorical variables were presented as the frequency (% percentage). The performance of vitamin D level was assessed using receiver operating characteristic (ROC) curve analysis and by calculating the area under the curve (AUC). The vitamin D level with the closest sensitivity and specificity value was chosen as the cut-off value. A P-value < 0.05 was considered significant. SPSS statistical software version 25 was used for analyses.
4. Results
Out of 200 patients in the study, 123 (61.5%) were female, and 77 (38.5%) were male; the patients had a mean age of 46.9 ± 14.9 years. Age and gender distributions, comorbid diseases, Fok 1 and Taq 1 polymorphisms, and vitamin D levels are shown in Table 1. The distribution of comorbid diseases is shown in Table 2.
| COVID-19 Positive | COVID-19 Negative | Totally | P-Value | |
|---|---|---|---|---|
| Age, y (mean ± SD) | 48.4 ± 15 | 45.4 ± 14.8 | 46.9 ± 14.9 | 0.163 b |
| Female | 44 (44.0) | 79 (79.0) | 123 (61.5) | <0.001 c |
| Male | 56 (56.0) | 21 (21.0) | 77 (38.5 ) | |
| Type 2 DM | 23 (23.0) | 2 (2.0) | 25 (12.5) | <0.001 b |
| Hypertension | 29 (29.0) | 9 (9.0) | 38 (19.0) | <0.001 b |
| Any comorbid disease | 40 (40.0) | 10 (10.0) | 50 (25.0) | <0.001 b |
| fok1 G/G | 52 (52.0) | 51 (51.5) | 103 (51.8) | 0.548 c |
| G/A | 42 (42.0) | 38 (38.4) | 80 (40.2) | |
| A/A | 6 (6.0) | 10 (10.1) | 16 (8.0) | |
| Taq1 G/G | 12 (12.0) | 17 (17.2) | 29 (14.6) | 0.098 c |
| G/A | 46 (46.0) | 31 (31.3) | 77 (38.7) | |
| A/A | 42 (42.0) | 51 (51.5) | 93 (46.7) | |
| Vitamin D level, ng/mL (mean ± SD) | 16.2 ± 11.3 | 26.7 ± 15.9 | 21.4 ± 14.7 | <0.001 b |
a Values are expressed as No. (%) unless otherwise indicated.
b Student's T test
c Chi-square tests
| Comorbid Disease | COVID-19 Positive (n:100) | COVID-19 Negative (n:100) | Totally |
|---|---|---|---|
| Hypertension | 29 | 9 | 38 |
| Type 1 DM | 23 | 2 | 25 |
| Asthma bronchiale | 9 | - | 9 |
| Ischemic heart disease | 6 | - | 6 |
a These differences between the groups were statistically significant (P < 0.001).
In all participating patients, the genotypic frequencies of Fok1 were 51.8% G/G, 40.2% G/A, and 8.0% A/A. Among COVID-19-positive patients, the frequencies were 52.0% G/G, 42.0% G/A, and 6.0% A/A. For the COVID-19 negative control group, the frequencies were 51.5% G/G, 38.4% G/A, and 10.1% A/A. There was no significant difference in Fok1 genotypic frequencies between the COVID-19 positive and negative control groups (P = 0.548). In all participating patients, the genotypic frequencies of Taq1 were 14.6% G/G, 38.7% G/A, and 46.7% A/A. Among COVID-19-positive patients, the frequencies were 12.0% G/G, 46.0% G/A, and 42.0% A/A. For the COVID-19 negative control group, the frequencies were 17.2% G/G, 31.3% G/A, and 51.5% A/A. There was no significant difference in Taq1 genotypic frequencies between the COVID-19 positive and negative control groups (P = 0.098). In the study, 200 people with COVID-19 positive and negative were included, and the mean vitamin D level for both groups was 21.4 ± 14.7 ng/mL. The COVID-19 positive group had a mean vitamin D level of 16.2 ± 11.3 ng/mL, while the COVID-19 negative group had a mean level of 26.7 ± 15.9 ng/mL. The difference between the two groups was statistically significant (P < 0.001).
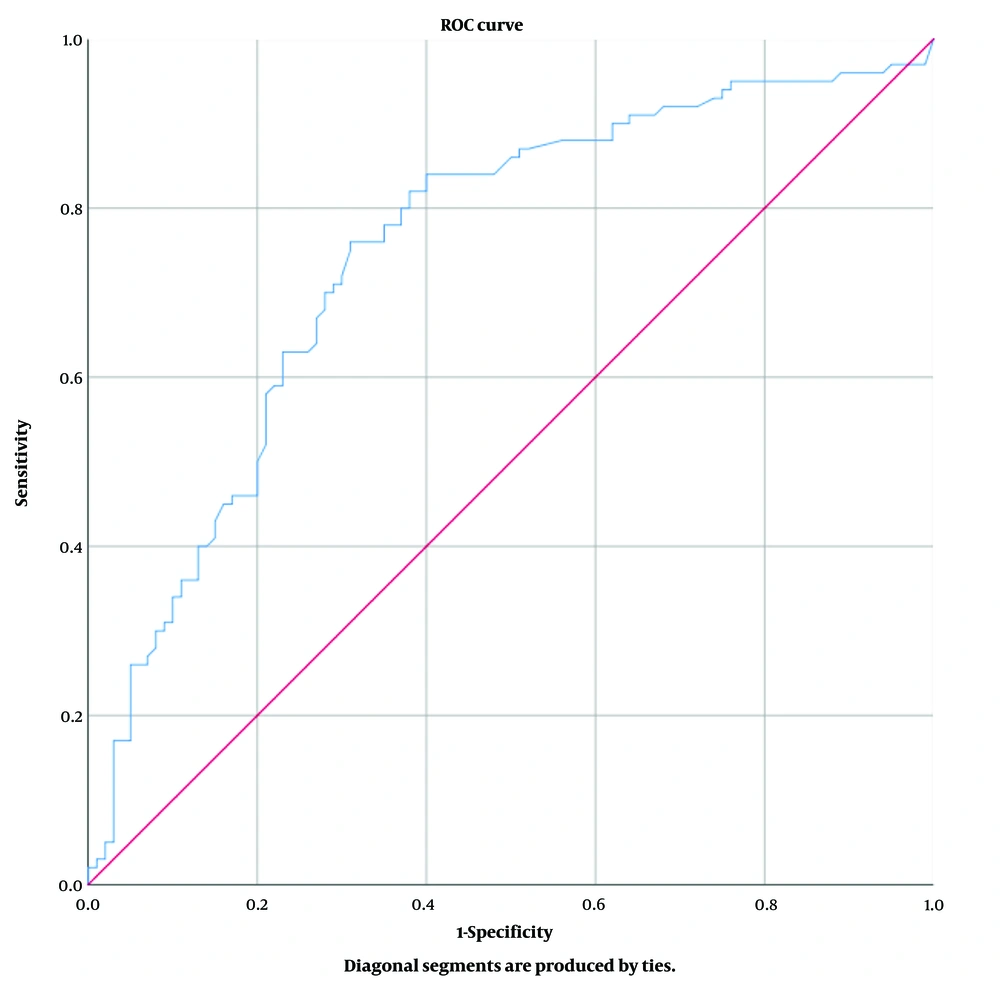
The effect of Vitamin D level on COVID-19 positivity was evaluated by performing ROC analysis (Figure 1). Our study found that the area under the curve for the ROC analysis was 0.748 (with a 95% confidence interval for the lower and upper bounds of 0.679 to 0.817), a sensitivity and specificity of 71%, and a significant cut-off value of 18.4 for predicting COVID-19 positivity in relation to vitamin D levels (P < 0.001). The odds ratio for the risk of COVID-19 positivity was calculated based on vitamin D levels. The analysis showed that the risk of COVID-19 positivity increased by 2.448 times (95% confidence interval: 1.758-3.410) for those with vitamin D levels below 18.4, compared to those with higher levels (P < 0.001) (Figure 2).
This study analyzed 82 COVID-19-positive patients who underwent thoracic CT. Out of the total, 7 patients exhibited no signs of lung involvement, while 75 did. The CT scans of patients with lung involvement exhibited images related to COVID-19 pneumonia, including ground-glass opacities, consolidation, and reticular patterns. The statistical analysis, using the Mann-Whitney U test, did not reveal any significant difference in median age, vitamin D levels, or Fok 1 and Taq 1 polymorphisms between patients with and without lung involvement (P = 0.584 and P = 0.921, respectively). Results are presented as median (25 - 75) percentile. A comparison of Fok1 and Taq1 polymorphisms and lung involvement is shown in Table 3.
| No Signs on Thoracic CT (N = 7) | Positive Signs on Thoracic CT (N = 75) | TOTAL (N = 82) | P-Value | |
|---|---|---|---|---|
| Fok1 pol | 0.733 | |||
| G/G | 3 (42.9) | 37 (49.3) | 40 (48.8) | |
| G/A | 4 (57.1) | 34 (45.3) | 38 (46.3) | |
| A/A | 0 (0.0) | 4 (5.3) | 4 (4.9) | |
| Taq1 pol | 0.768 | |||
| G/G | 1 (14.3) | 9 (12.0) | 10 (12.2) | |
| G/A | 4 (57.1) | 34 (45.3) | 38 (46.3) | |
| A/A | 2 (28.6) | 32 (42.7) | 34 (41.5) |
a Chi-square tests were used. Values are expressed as No. (%).
5. Discussion
In our study, it was found that COVID-19-positive patients had significantly lower mean vitamin D levels and a higher prevalence of severe vitamin D deficiency compared to the COVID-19-negative control group. When evaluating other studies investigating the relationship between vitamin D levels and COVID-19, a study involving 204 COVID-19 patients found a mean vitamin D level of 12 ng/mL and a 41.7% prevalence of vitamin D deficiency (13). Ilie et al. also investigated COVID-19 cases per million population and vitamin D levels in various countries. They found a significant relationship between vitamin D levels and the number of COVID-19 cases in different countries (14). Calcitriol, the active form of vitamin D, can reduce SARS-CoV-2 titers and inhibit viral replication in nasal epithelial cells, leading to decreased viral load in patients with sufficient vitamin D levels. The meta-analysis of 8209 patients found that there is a 1.5 times increase in COVID-19 positivity when serum vitamin D levels are below 30ng/mL (15). Additionally, the meta-analysis of 31 observational studies and a systematic review determined that COVID-19-positive patients had a mean vitamin D level that was 5.9 ng/mL lower than COVID-19-negative patients (1, 15-19).
We calculated the odds ratio and found that the risk of COVID-19 positivity increased approximately 2.5 times if the vitamin D level was below 18.4 ng/mL. This means that a cut-off value of 18.4 ng/mL was determined for COVID-19 susceptibility. Since our patients were hospitalized COVID-19 patients, this value can be a determining factor for hospitalization. Similarly, Radujkovic et al. reported that a vitamin D level below 20 ng/mL was significant for hospitalization (20). A meta-analysis of 14 studies involving 999,179 patients investigating low serum 25(OH) Vitamin D levels found that low serum vitamin D levels were associated with high rates of COVID-19 (21). Another study investigated the relationship between Vitamin D levels and lung infection in 6,789 patients with COVID-19. It showed that for every 4 ng/mL increase in vitamin D levels, the probability of lung infection in COVID-19 patients decreased by 7% (22).
Comorbidities such as type 2 diabetes and hypertension may increase susceptibility to COVID-19. Our study found a significantly higher prevalence of these chronic diseases in the COVID-19-positive group compared to the negative control group. Other studies have also reported a higher prevalence of chronic diseases among COVID-19 patients than among negative patients (23, 24). It has been proven that polymorphism of the gene encoding vitamin D receptor synthesis is associated with hypovitaminosis D (25). VDR gene polymorphism affects VDR activity by generating a dysfunctional receptor. Therefore, vitamin D receptor gene polymorphisms may affect the effector response against SARS-CoV-2.
Our study investigated the relationship between VDR gene polymorphism and susceptibility to COVID-19 infection. Fok 1 and Taq 1 polymorphisms, which affect VDR activity, were compared between the patient and control groups, but no statistically significant difference was found. Our study found no significant differences in vitamin D levels and VDR gene polymorphisms between patients with and without lung involvement. While Abdollahzadeh et al. also found no significant relationship between Taq1 polymorphism and COVID-19 susceptibility in their study of 500 patients, a recent study of a Cuban population found that Taq1 polymorphism may increase susceptibility to COVID-19 (26, 27). These divergent results suggest that ethnicity and geography may influence the relationship between Taq1 polymorphism and COVID-19 susceptibility.
Batur conducted a study across 26 countries on the prevalence and mortality rates of COVID-19 and VDR gene polymorphisms (28). The study identified gene polymorphisms in Taq1, Fok1, Apal, and Bsml loci and found a significant relationship between Apal and Taq1 polymorphisms and recorded the prevalence and mortality rates of COVID-19 December 6, 2020, suggesting that VDR gene variations in different nationalities may explain variations in the prevalence of COVID-19. However, similar to our study, no relationship was found between Fok1 polymorphism and the prevalence of COVID-19. The study by Zeidan et al. from Egypt found that the FF genotype of Fok1 was significantly higher in COVID-19 patients, and patients with this genotype showed a higher susceptibility to COVID-19 infection (29). Ethnic group differences could explain discrepancies between the study results.
5.1. Conclusions
Vitamin D has immunoregulatory properties that can protect against COVID-19 through various mechanisms, and our study found a correlation between low vitamin D levels and increased prevalence of COVID-19. The study established a cut-off value of 18.4 ng/mL for vitamin D, with levels above this value being protective against COVID-19. In contrast, levels below this value increased the risk of COVID-19 positivity 2.5 times. While gene polymorphisms in the vitamin D receptor can cause D hypovitaminosis, potentially leading to increased susceptibility to COVID-19, our study found no association between Fok 1 and Taq 1 polymorphisms and increased prevalence of COVID-19. Our study suggests that comorbidities such as diabetes and hypertension may be risk factors for the development of COVID-19.