1. Background
In the last decade, the pattern of antibiotic resistance through integrons in Pseudomonas aeruginosa has been increasing. Integron P. aeruginosa is an opportunistic pathogen that has a strong potential to cause severe nosocomial infections and serious difficulty in burn patients. This organism shows notable antimicrobial resistance and is often resistant to multiple antibiotics. Integron genes as mobile genetic elements play a central role in the spread of P. aeruginosa antibiotic resistance. The rapid transmission and spread of these organisms, which can produce the aforementioned enzymes, has increased the rate of hospital infection around the world. Furthermore, due to the resistance of these microorganisms to a wide variety of antibiotics recently, the therapeutic strategy for these types of infections caused by them has been difficult and has led to increased mortality. The difficulty of eradicating P. aeruginosa infection is due to its intrinsic resistance to different antibiotics caused by several mechanisms, including low outer membrane permeability, overexpression of efflux pump system, and enzymatic antibiotic modifications, e.g., β-lactamase yield.
Due to the resistance of this pathogen to a wide range of antibiotics, it has become difficult to treat infections caused by it in recent years. The mechanisms of bacterial resistance to various antibiotics vary. One such mechanism involves the production of metallo-β-lactamase enzymes, which are encoded by different genes such as Imp-1, Imp-2, Vim-1, and Vim-2. These enzymes are a contributing factor to antibiotic resistance (1).
Metallo-β-lactamases are enzymes produced by non-fermenting Gram-negative bacilli such as P. aeruginosa. The strains that produce these enzymes are more resistant to beta-lactam antibiotics such as cephalosporins, penicillins, and carbapenems (2, 3). These metallo-β-lactamases have various types, including Imp, Vim, Spm, Gim, Aim, Sim, and Uim, and are divided into four categories A, B, C, and D based on their molecular structure. Group A hydrolyzes penicillin and cephalosporins. Type B, which depends on zinc metal, is capable of hydrolyzing carbapenem antibiotics and is divided into three subclasses (B1, B2, and B3). Type C or AMPc can hydrolyze cephalosporins and neomycin, and type D can hydrolyze Cloxacillin and oxacillin (4).
The genes encoding these enzymes are both plasmid and chromosomal, which are easily transferred to other bacteria. In P. aeruginosa, the most important genes are Vim and Imp, which are among the metallo-β-lactamase plasmid genes. The resistance to the carbapenem group antibiotics (imipenem) is due to the presence of these genes, which can be easily transferred to other strains (5). Reports show that Pseudomonas strains producing these metallo-β-lactamases can increase mortality in patients. The genes encoding these enzymes are either of chromosomal origin or genetic elements such as plasmids, transposons, and integrons (6). In the last decade, the pattern of antibiotic resistance through integrons in P. aeruginosa has been increasing.
2. Objectives
This study evaluated the abundance of integrons and the pattern of resistance to carbapenems (metallo-β-lactamases including Spm, Imp, and Vim) and its correlation with the presence of different classes of integrons (classes I and II) in P. aeruginosa isolates from burn patients.
3. Methods
3.1. Research Population
In this descriptive-analytical cross-sectional study, the population included patients with burn infections referred to the burn center of Velayat Hospital, Rasht city, Iran.
3.2. Collection, Preparation, and Identification of Samples and Maintenance of Isolates
This study was conducted on 73 samples of P. aeruginosa isolated from the burn wounds of patients admitted to a burn center. Samples were taken from the burn site using a swab. The swab soaked with the sample was inoculated in a TSB medium. To confirm the phenotype of P. aeruginosa, Gram staining and diagnostic biochemical tests, including oxidation-fermentation (OF) test, production of pigments, citrate, catalase, oxidase, and growth at 42°C were used. The standard strain of P. aeruginosa ATCC 27853 was used as a control.
3.3. Molecular Identification of Bacteria
Extraction of bacterial DNA using the boiling method: The samples were cultured on Mueller Hinton culture medium and incubated for 24 hours at 37°C. After ensuring the growth, some colonies were removed from the surface of the plate and dissolved in 1.5 mL of TE buffer solution in a microtube and centrifuged at 4 000 rpm for 5 min. Then, the supernatant was discarded, and 200 TE was added to the bottom of the microtube and kept at room temperature for 10 minutes. One hundred degrees Celsius in a hot plate machine (dry bath, Taiwan). After completing this step, they were centrifuged at 14 000 rpm for 10 min. The supernatant was collected in a sterile Eppendorf tube. A biospectroscopy device (Thermo Scientific, UK) was used to measure DNA concentration.
Measuring the concentration of extracted DNA: To measure the concentration of extracted DNA, the optical absorption of the sample was used in a UV bio-spectrophotometry device (Thermo Scientific, UK). It was diluted and transferred to the cuvette, and the approximate concentration of the sample was calculated by the device. To ensure the purity of the extracted DNA, we divided the absorbance of the wavelength of 260 nm by the absorbance of the wavelength of 280 nm, and the resulting value should be about 8.1 μg/μL (7).
Polymerase chain reaction: In this study, the primers related to all the genes, including IntI1, IntI2, Spm, vim, and imp, were designed using Oligo Primer Analysis Software Version 7. The nucleotide sequence of the primers is shown in Table 1 (7). A mixture of reaction materials (Master Mix) was prepared, and after adding template DNA, it was placed in a thermocycler (SimpliAmp, UK) according to the temperature program. Conducting polymerase chain reaction for plasmid carbapenemase class A, B, and D beta-lactamase genes: In this step, the strains that were identified for disk imipenem by disk diffusion method were selected to check the plasmid genes of carbapenemase using the polymerase chain reaction (PCR) method.
| Primer | Sequence |
|---|---|
| blaSpm-F | AAAATCTGGGTACGCAAACG |
| blaSpm-R | ACATTATCCGCTGGAACAGG |
Amount and Type of Polymerase Chain Reaction Materials (the Same Concentrations Were Used for Other Polymerase Chain Reactions)
Identification of beta-lactamase plasmid genes (blaImp and blaVim) using PCR technique: The steps were done using a Pantaplex PCR device. The PCR was carried out in a volume of 50 μL, including 48 μL of master mix and 4 μL of DNA with 10 picomol gel. The master mix contained 2.5 μL of 10X buffer, 2.25 µM MgCl, 200 µM dNTPs, and 0.3 μL of 500 unit single polymerase enzyme.
Identification of carbapenemase blaSpm plasmid gene by PCR: The PCR was performed in a volume of 25 microliters, including 15 μL of master mix and 4 μL of DNA with 10 picomol gel. The master mix contained 2.5 μL of 10X buffer, 2.25 μM MgCl, 200 μM dNTPs, and 0.3 μL of single polymerase enzyme of 500 units (Tables 2 and 3).
| Primer | Sequences |
|---|---|
| IntI1 | GGTCAAGGATCTGGATTTCG |
| ACATGCGTGTAAATCATCGTC | |
| IntI2 | CACGGATATGCGACAAAAAGGT |
| GTAGCAAACGAGTGACGAAATG |
Primers to Identify SPM Metallo-Beta-Lactamases
| Primer | Sequence |
|---|---|
| blimp-F | GGAATAGAGTGGCTTAAYTCTC |
| blimp-R | GGTTTAAYAAAACAACCACC |
| blaVim-F | GATGGTGTTTGGTCGCATA |
| blaVim-R | CGAATGCGCAGCACCAG |
Primers for Identifying Integrons
3.4. Electrophoresis
3.4.1. (A) Preparation of Agarose Gel
Based on DNA length, 1.5% gel was used, and 1.5 g of agarose powder was dissolved in ML100 TBE buffer and heated in the microwave for further dissolution until a clear and uniform liquid was obtained (Figure 1).
3.5. Data Analysis Method
To check the relationship between the data obtained in different stages of this research, the data were entered into SPSS statistical software and analyzed. The chi-square test was used to determine the relationship between groups.
4. Results
4.1. Sampling and Gender
The results from 73 patients showed varying frequencies and percentages of gender distribution, with 60 samples from males (82.2%) and 13 samples from females (17.8%) (Table 4) (Figure 2).
| Gender | Frequency (%) |
|---|---|
| Man | 60 (82.2) |
| Female | 13 (17.8) |
| Total | 73 (100) |
Frequency of Gender
4.2. Distribution of Isolates Based on the Type of Infection
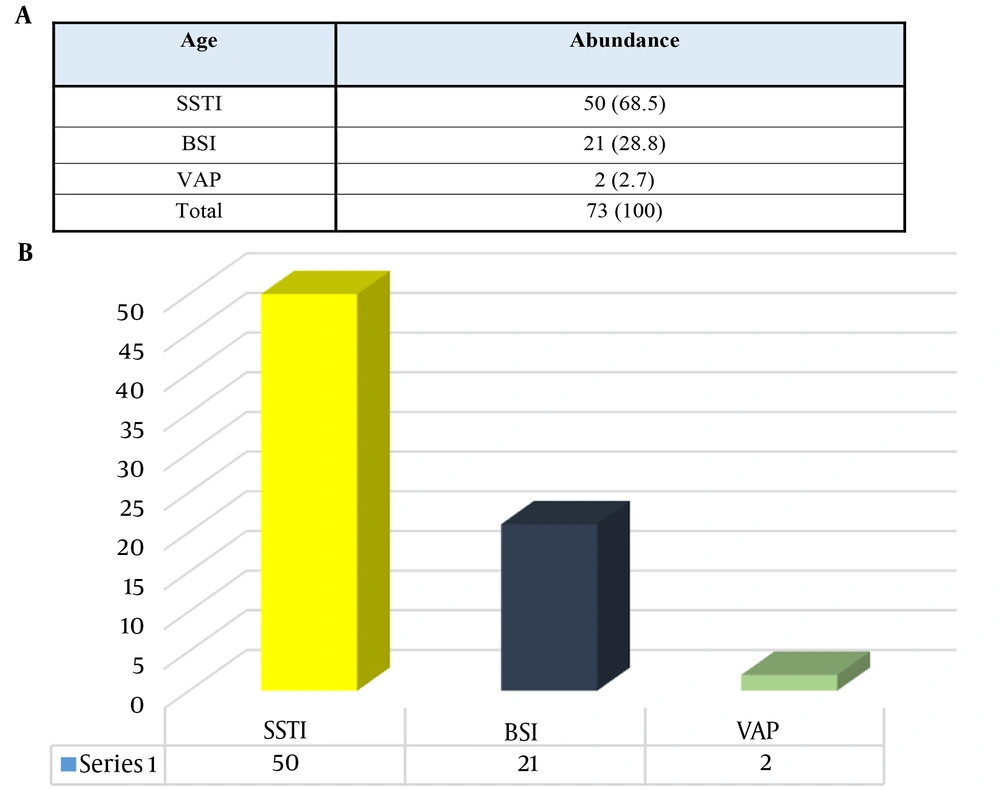
Based on the results, patients were generally divided into 3 categories, including those with skin and soft tissue infection (SSTI), bloodstream infection (BSI), or ventilator-induced pneumonia (VAP) samples. The SSTI samples, with 68.5%, were the most frequent, and the lowest frequency was related to the VAP samples, with 2.7% (Figure 3).
4.3. Frequency Integrons
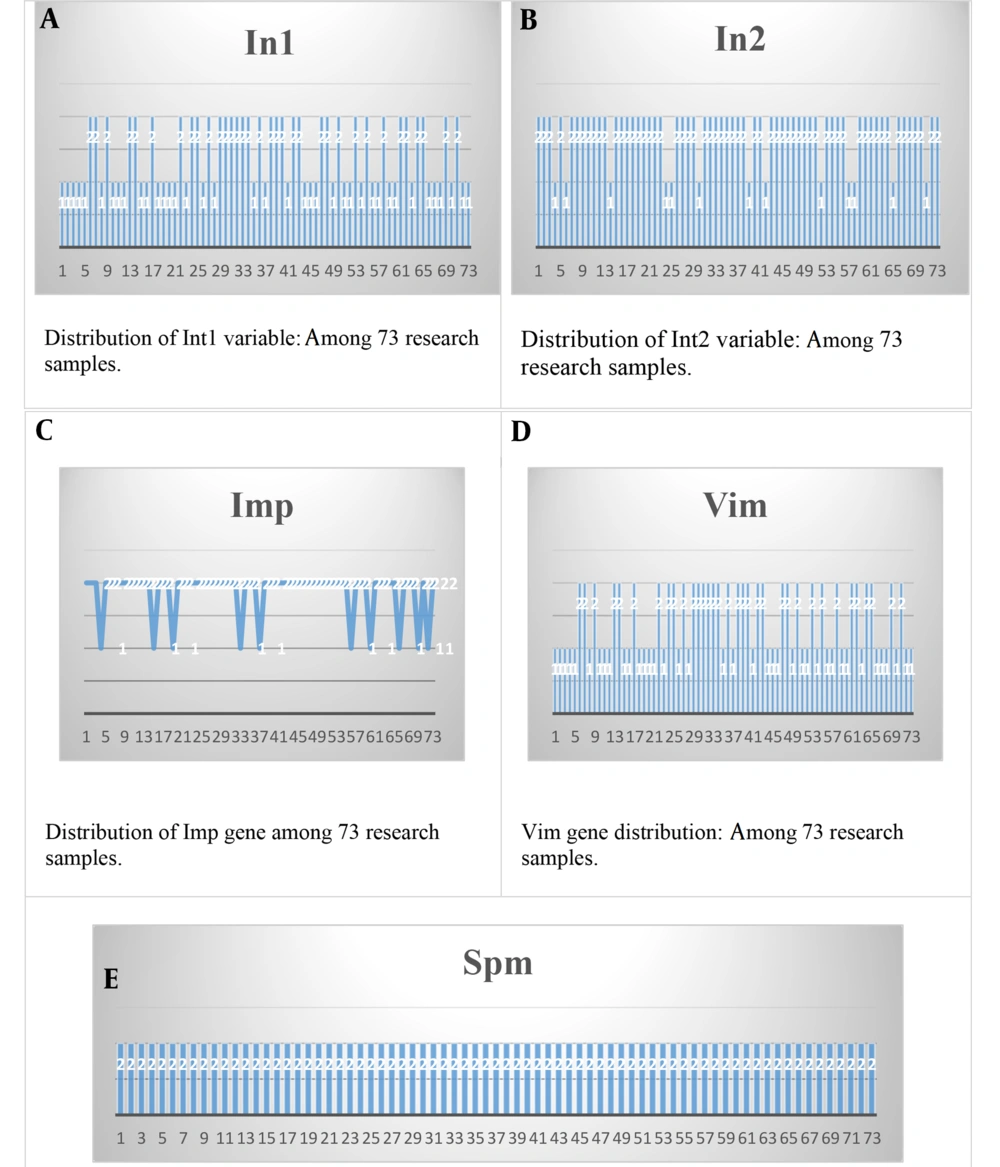
Table 5 shows the frequency and percentage of integron I. The results showed that 39 people (53.4%) had a positive reaction, and 34 people (46.6%) had a negative reaction. Also, the results of the frequency of the index integron 2 showed that 13 people (17.8%) had a positive reaction, and 60 people (82.2%) had a negative reaction.
| Abundance (%) | |
|---|---|
| Integron 1 | |
| Positive | 39 (53.4) |
| Negative | 34 (46.6) |
| Total | 73 (100) |
| Integron 2 | |
| Positive | 13 (17.8) |
| Negative | 60 (82.2) |
| Total | 73 (100) |
Frequency Integrons
4.4. Frequency Genes
Table 6, concerning the frequency and frequency percentage of the Imp gene, shows that 10 bacterial isolates (13.7%) had a positive reaction, and 63 bacterial isolates (86.3%) had a negative reaction. Also, the results of Vim gene frequency showed that 16 bacterial isolates (21.9%) had a positive reaction, and 57 bacterial isolates (78.1%) had a negative reaction.
| Abundance (%) | |
|---|---|
| Imp | |
| Positive | 10 (13.7) |
| Negative | 63 (86.3) |
| Total | 73 (100) |
| Vim | |
| Positive | 16 (21.9) |
| Negative | 57 (78.1) |
| Total | 73 (100) |
| Spm | |
| Positive | 0 (0) |
| Negative | 73 (100) |
| Total | 73 (100) |
Gene Frequencies
4.5. Analytical Findings
Distribution of clinical samples and different departments based on class I and II integron-producing isolates (Table 7) showed that 58.98% of isolates carrying class I integron were isolated from the intensive care unit.
| Cases | Integron-1, Positive, N = 39 | Integron-2, Positive, N = 13 |
|---|---|---|
| Burn accidents | 16 (41.02) | 5 (38.4) |
| Intensive care | 23 (58.98) | 8 (61.6) |
| Clinical samples | ||
| SST | 27 (69.2) | 8 (61.5) |
| BSI | 10 (25.6) | 4 (30.8) |
| VAP | 2 (5.2) | 1 (7.7) |
Distribution of Clinical Samples and Different Departments Based on Class I and II Integron-Producing Isolates a
4.6. Examining the Relationship Between Type I Integron and Triple Genes
Considering that the value of significance was less than the level of 0.05 (Sig. = 0.012), there was a significant relationship between type I integron and the Imp gene, and the intensity of this relationship was 0.292. Considering that the correlation test between triple genes and two types of integron I and II was significant only in the case of IMP gene and integron type I and considering the positive value of correlation (0.292), it can be concluded that with the increase of the value of integron 1, the intensity of IMP gene transfer increased (Table 8).
Examining the Relationship Between Type I Integron and Triple Genes
4.7. Examining the Relationship Between Integron Type II and Triple Genes
Considering that the value of significance exceeded the level of 0.05 (Sig. = 0.494), there was no relationship between type II integron and the Imp gene. Also, considering that the value of significance exceeded the level of 0.05 (Sig. = 0.115), there was no relationship between type II integron and the Vim gene (Table 9).
Examining the Relationship Between Integron Type II and Triple Genes
5. Discussion
Pseudomonas aeruginosa is one of the main pathogens involved in hospital infections, which is very important in clinical samples, including burns and wounds (8). In many reports from burn centers, this microorganism is considered the most common bacterial species isolated from all types of burn wounds (9). The present study investigated the prevalence of P. aeruginosa and its mechanism of carbapenem resistance in burn patients at Rasht Province Hospital, as P. aeruginosa isolates can cause various wound and burn infections. Based on the survey results regarding the prevalence of P. aeruginosa in various study groups, it was found that this bacterial pathogen was present in over 82% of men and nearly 18% of women, as well as in related clinical samples. In this regard, the frequency of clinical strains of P. aeruginosa isolated from different departments of Tehran hospitals was investigated, and it was reported that 70% of isolates were found in men (10). Also, a related study reported that 66.61% of the 60 clinical samples of P. aeruginosa collected from Tehran's Di and Motahari hospitals belonged to men, and the other 34.38% belonged to female patients with related infection symptoms.
The consistency of the results reported in our study and other similar studies indicates that this bacterium is more prevalent in men and in individuals who are more susceptible to this infectious pathogen in various wound and burn complications. This is due to the higher incidence of severe cases in men, leading to more hospitalizations compared to women. Pseudomonas aeruginosa in patients hospitalized in the intensive care unit of the hospital has a high importance in pathogenicity because it includes a high percentage of isolates and has caused death in this unit (11). In this way, VAP is one of the most common hospital infections associated with P. aeruginosa in the intensive care unit, and its rate has been reported up to 28% in those who have had a history of using artificial respiration devices (12). In our study, we examined the majority of Pseudomonas isolates obtained from burn patients who were exclusively hospitalized in the hospital's special care and burn accident departments. The results indicated a frequency of nearly 55% and more than 45% of this bacterium, respectively.
The section mentioned above indicated that these patients often suffer from immune system deficiency due to an underlying disease. Actions such as invasive medical devices, such as intravenous catheters and ventilators, are the reason for increasing the infection rate in these patients (13). Another study examined the molecular epidemiology of P. aeruginosa, finding that 47.5% of the samples were attributed to prolonged stays in the intensive care unit (ICU) (14). In the present study, the analysis of Pseudomonas isolates obtained from individuals with burns revealed that over 68% of cases were associated with SSTIs, a common occurrence in burn and wound cases. In a similar study, Morris and Cerceo reported wound infections with more than 33% as the most common cases of the presence of P. aeruginosa. In that study, the lowest prevalence was related to blood infection. Often, if P. aeruginosa becomes resistant to one class of antibiotics, it can become resistant to other classes as well, which leads to the emergence of multi-drug resistant (MDR) strains (15).
To deal with isolates with multiple resistance, carbapenems were considered one of the most appropriate drugs to treat infections caused by gram-negative bacteria such as P. aeruginosa. Among beta-lactam antibiotics, carbapenems have the widest spectrum of antibiotic activity and are the strongest agent, which has been mentioned in various reports. Imipenem and meropenem are traditionally used in the treatment of hospital infections. However, many studies have been conducted on the resistance of P. aeruginosa isolates to carbapenems, and different results have been reported in different parts of the world (16). Importantly, we examined and tested the presence of carbapenem resistance genes, specifically metallo-beta-lactamases Imp, Vim, and Spm, as well as genes intI1 and intI2, which are associated with the presence of integron factors. The results indicated the presence of all target genes in P. aeruginosa isolates except for the Spm gene, which was not detected in any of the isolates. The results indicated the presence of all target genes in P. aeruginosa isolates except for the Spm gene, which was not detected in any of the isolates.
Metallo-β-lactamase genes are located in plasmids or integrons and, therefore, can transfer to other bacteria (17). Metallo-β-lactamase genes have five groups (Vim, Imp, Spm, Sim, and Gim). There are different variants of Imp and Vim genes that have a global distribution, while some of these genes, such as Spm, have been found only in specific regions (18). In our study, apart from the Imp and Vim genes, which have global prevalence, the Spm gene, which has a rapid spread and high mortality in epidemics, was also investigated. Based on the obtained results, 13.7% of the samples had the Imp gene, and the other 21.9% had the Vim gene in their genetic material, which indicates a high level of metallo-β-lactamase genes encoding resistance to antibiotics such as carbapenems. It was also mentioned that the Spm gene was not observed in any of the examined isolates. Various studies have been conducted on the relative frequency of these genes in Pseudomonas clinical samples, yielding different results and reports. Each study is in some way related to the significance of antibiotic resistance in bacterial isolates and the potential transfer of this microbial characteristic, which can lead to medical problems.
In a study, 20 Vim metallo-β-lactamase genes in P. aeruginosa isolates under investigation in burn patients were detected. In their reports, no isolates containing the Imp gene or other genes involved in the occurrence and transmission of antibiotic resistance were observed (19). Sedighi et al. investigated 68 clinical isolates of P. aeruginosa resistant to imipenem to detect Spm, Vim, and Imp metallo-β-lactamase genes. Their results indicated the detection of 16 isolates producing MBL, all of which were related to the Vim gene, and none of them had the Imp or Spm gene (20). In a study, 75 metallo-β-lactamases-producing samples were identified, of which 70 Vim isolates (33%) and 20 Imp isolates (9%) were positive (21). In another study, Moosavian and Rahimzadeh (as cited by Vural et al.) isolated 236 clinical isolates of P. aeruginosa from different parts of the body, of which 110 isolates were positive by the MBL phenotypic method. Then, among the isolates producing metallo-β-lactamases by a phenotypic method using molecular methods, 55% and 1.6% were reported to have Imp and Vim genes, respectively (22).
Generally, the horizontal transfer of resistance genes is considered a major cause of facilitating the rapid spread of antibiotic resistance in microorganisms. Our results show the importance of class I and II integrons in antibiotic resistance and its relationship with P. aeruginosa isolates with carbapenem resistance genes. Therefore, it can be acknowledged that integrons play an important role in the acquisition and dissemination of antibiotic resistance genes among these pathogens, so the management of infection control and the appropriate use of antibiotics will be necessary to control the dissemination of antibiotic resistance genes.
5.1. Conclusions
The variable frequency of classes I and II integron genes has been confirmed in different studies. It is also worth noting that class I integron may be present in isolates that are sensitive to one type of antibiotic and resistant to another. Therefore, it can be assumed that antibiotic-resistant integron genes can contain one or more types of resistance genes in their structure, which cause resistance to that particular type of antibiotic. There was a statistically significant relationship between class I integron and the blaImp gene, but in the rest, no such relationship was detected. In addition, a bacterial isolate lacking integrons but resistant to one or more types of antibiotics indicates that mechanisms other than integrons play a role in resistance and the presence of related genes. Importantly, all issues related to antibiotic resistance should be considered in future studies.