1. Background
Of the infectious agents that are transmitted through food, the ones with the largest share are Salmonella species. The transmission pathways of Salmonella spp. isolates to humans include contaminated food, undercooked meat, and eggs (1). Salmonella enteritidis are bacteria that have been known for 125 years, and have maintained their importance as an infectious agent in humans. The U.S. FoodNet surveillance program reported that Salmonella serotypes were the second leading cause of bacterial food-borne infections in 2004, with 5 serotypes accounting for 59% of the Salmonella infections (2). The serotypes called Typhimurium, Enteritidis, and Newport are the most common. In the recent years, approximately 1.5 million gastroenteritis cases in the USA were caused by S. enterica serovars. Widespread infection is still an important public health issue (3).
Reliable methods should be preferred for accurate epidemiological analysis of salmonellosis outbreaks. Among the microbiological analyzes, the most preferred methods are culture tests, biochemical tests, serotype determination, and phage type. During the last 2 decades, the recommended surveillance techniques have been DNA based (4). As a molecular method, the pulsed-field gel electrophoresis (PFGE) method, which has been preferred by CDC since the early 1990s, remains the gold standard (5-7).
It is important for all phases to be under control in the process of preparing food. All stages, starting from live animals, especially farm products, to the cooking stage must be perfomed with caution. Surveillance programs to monitor contamination with Salmonella must include the whole food chain at regular intervals for preventing food-borne Salmonella infections (8). Sanitary measures for preventing Salmonella contamination necessitates obtaining uncontaminated feeds, controlling rodents, and wild birds, while restricting the flow of personnel, equipment, and air to “clean” areas, maintaining the sanitation of transport vehicles. In addition, Salmonella in feeds may be eliminated by thermal and chemical processing (9).
2. Objectives
In the current study, the researchers investigated food associated S. enteritidis outbreak in factory workers of Turkey.
3. Methods
Medical treatment was sought by 257 ill individuals, who were kept in the emergency area for observation. Among 257 patients with diarrhea, abdominal pain, headache, fever and vomiting, 48 (19%) were hospitalized. All cases were male. Stool samples were plated on Salmonella-Shigella agar (Salubris, Turkey), Eosine Methylene Blue agar (Salubris, Turkey), and 5% sheep blood agar (Salubris, Turkey). Plates were incubated at 37°C for 18 to 24 hours. Suspected colonies were selected for further identification on the next day, using API 20E (BioMerieux, France). Isolates were sent to reference laboratories for further serotyping. Conventional serotyping was conducted at the national enteric pathogens reference laboratory, public health institution of Turkey under the national enteric pathogen laboratory surveillance Network (EPLA). In the Reference Laboratory, through the Kauffmann-White scheme, O (Somatic), Vi (capsular), and H (flajella) antigens (Public Health Institution, Turkey) were formulated and the serotype was assigned.
Pulsed field gel electrophoresis was used for the epidemiological analysis of isolates and PFGE analysis was performed by XbaI macrorestriction enzyme, according to the world health organization protocols (10). Antimicrobial susceptibility was determined by disc diffusion method as recommended by CLSI (11). Mueller-Hinton agar (Salubris, Turkey) was used for all strains. The antibiotic discs included ampicillin, chloramphenicol, kanamycin, tetracycline, gentamicin, trimethoprim-sulfamethoxazole, nalidixic acid, ceftazidime, cefotaxime, and ciprofloxacin (BD, Diagnostic Systems, USA). Escherichia coli ATCC 25922 was used as the control strain.
All food and water samples were also collected and investigated by the Public Health Institution of Izmir. Food samples (soup, meatball, etc.) were investigated for the detection of microbiological contamination level. The study was approved by the ethics committee of Izmir Ataturk Education and Research Hospital on the 21st of May, 2015 (code 101).
3.1. Ethics Statement
The study was approved by the ethics committee of Izmir Ataturk Education and Research Hospital on the 21st of May, 2015 (code 101).
4. Results
During the outbreak, 10 out of 48 (21%) stool samples and 1 out of 25 (4%) blood culture from patients were positive. All patients were male. Despite being rare, 1 Salmonella spp. isolate was isolated from the blood culture of a 46-year-old male patient. A predisposing disease was not detected.
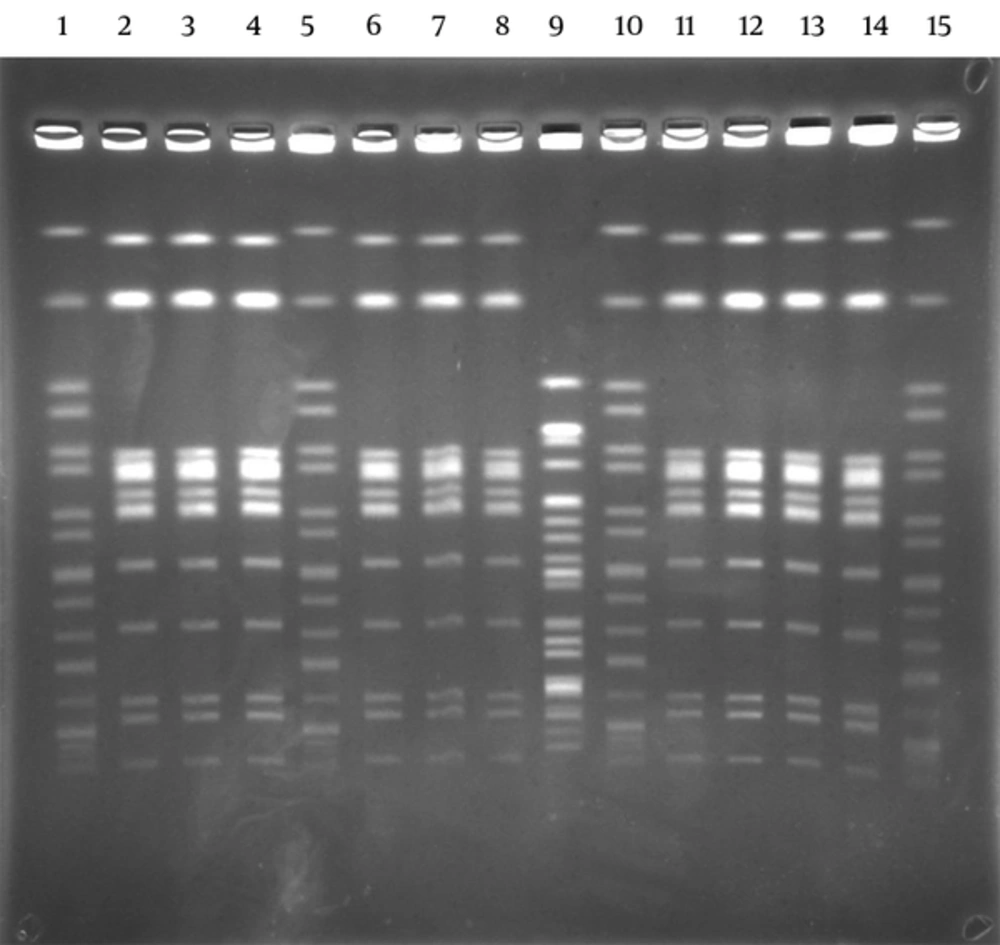
The lactose non-fermenting colonies on the Salmonella-Shigella agar plates were identified as S. enterica ssp enterica by API 20E (BioMerieux, France) in stool samples. Ten out of eleven strains were found in cases as S. enteritidis (9,12; g,m; -) serotype with the agglutination test. Outbreaks were caused by the same Salmonella serotype. By PFGE, 10 identical S. enteritidis patterns were shown with enzyme XbaI. Results showed that they all belonged to group D, S. enteritidis (9,12; g,m; -). One isolate showed a different pattern. This was considered as a sporadic case (Figure 1). All isolates (n = 11) were susceptible to ampicillin, nalidixic acid, ciprofloxacin, trimethoprim/sulfamethoxazole, tetracycline, kanamycin, chloramphenicol, gentamicin, ceftazidime, and cefotaxime. All patients were succesfully treated with ciprofloxacin 2 × 750 mg/day for 5 days and returned to work on the 7th day.
Food workers in the same factory had been monitored by culture of stool samples. Therefore, samples were collected and processed in a medical microbiology laboratory from food-handlers in the same factory. None of them were observed in the presence of Salmonellae. However, S. enteritidis was detected in one item of the food (meatball), which was consumed by the subjects by public health institution laboratory of Izmir. Results of the study showed that meatballs contaminated with S. enteritidis (9,12; g,m; -) were responsible for this outbreak. The same S. enteritidis strain was isolated from meatball and soup by the National Food Reference Laboratory/Turkey in this outbreak. This outbreak was concluded to have been caused by staff members of the factory, who contaminated the meatball during processing with eggs or meat products. Although meat products and eggs were kept under appropriate conditions for consumption in the factory kitchen, conditions of hygiene and sanitation were not considered susceptible enough from production till consumption.
5. Discussion
Salmonella enteritidis are bacteria that have been known for 125 years, and have maintained their importance as an infectious agent in humans. The first symptoms in the body after becoming infected are fever, stomachache, and diarrhea. The stated onset periods range between 12 and 72 hours. Infection becomes limited during 7 days, generally automatically or with antibiotic treatment. The course may be severe in elderly, children, and those with immune system failure. Therefore, in-service treatment in a hospital is advised as the infection that has started in the digestive system may spread to the entire body through the blood (12). Salmonella spp. infections could have mild or no symptoms, yet the pathogen could continue to be excreted from the body for up to 3 months after recovery (13).
About 400 people are known by CDC to die of acute salmonellosis in one year. Salmonella enteritidis is one of the most common serotypes of Salmonella reported worldwide (14). The number of outbreaks of S. enteritidis rose dramatically in the past 30 years (6). Salmonella enteritidis are the most commonly reported serotype (36%) in Turkey (15-17), as Salmonella spp. could survive in high temperatures, high fat, and low water content conditions (18). Contamination by direct contact occurs easily because they are present in the intestines of some mammals and poultry. Most of the infections were caused by contamination from poultry or egg products (19-21). In S. enteritidis infections, egg shell has an important role in the pathways of transmission through food. Infection develops after contaminated eggs are consumed. Other sources, such as raw milk, pork, beef, sprouts, and raw almonds are known less frequently. According to CDC data, travel-related Salmonella infections and carriage are seen after traveling to Asian and African countries, primarily Latin America. As reported previously 38% of all travel-related Salmonella cases reported by the CDC developed after travel to Mexico (22-24).
Overall, 176.395 Salmonella spp. cases in foodborne outbreaks until 2005 were reported in European Union countries. The most common source of outbreaks was eggs (25). Salmonella enteriditis and S. typhimurium are the most commonly reported serotype in this period. Salmonella enteritidis/S. typhimurium positivity rates were 20.4% in chicken and egg farms (26-29). In the USA, 1939 cases were reported in the epidemics from many regions from 1st of May to 30th of November, 2010, and the causing agent was S. enteritidis. These infections were associated with egg shells (6).
Again in 2014, 87 people were reported from 11 different states in S. enteritidis epidemics. In the course of an epidemic, 14 patients out of 52 (27%) had to be treated as in-patients. No death was reported (6). Local public health officials also performed traceback. The results of investigations on the source of these epidemics were related to bean sprouts. The strains isolated in antibiotic susceptibility test were reported to be susceptible. As of November, 2010, nine different survey protocols were prepared in the European Union -countries. Salmonella spp. analysis was included in these protocols in packing centers and eggs, which are sold in markets and shops (26). The control program was implemented in laying hens since 2008 in Turkey (30).
Oktem et al. Reported that 8 minutes of boiling was needed for the bacterium to become ineffective. It was pointed out in this study that Salmonella bacteria could still remain alive at the end of the 21st day at 4°C (31). It is necessary to avoid the consumption of undercooked egg and foods infected through touch with egg shells, especially for children, elderly and patients with immune system failure. Among protective precautions, washing hands with water and soap after contact with raw eggs is of great importance. Similarly, when there is a touch with surfaces, it is necessary to make the surface clean with chemical matters that provide special hygiene that could eradicate the bacteria. It is necessary for raw eggs to not be held at room temperature in order to reduce the risk factor on this issue. Eggs with dirty or cracked shells are not advised.
Pulsed field gel electrophoresis is commonly used to detect and identify the epidemic relationship between pathogens and meats. Each bacterial species was known by its unique DNA structure. The method, known as PFGE, is used to achieve this unique fingerprint (32-34). Ashtiani et al. (35) detected that ampicillin, amikacin, and chloramphenicol resistance among Salmonella spp. had increased between 1996 and 2005. There has been no significant change in cephalothin, trimethoprim-sulfamethoxazole, ceftazidime, and gentamicin resistance in this study.
Kesli et al. (36) reported that ampicillin resistance was determined as 63%. In addition, chloramphenicol 61%, cefuroxime 56%, ceftriaxone 49%, tetracycline 8%, trimethoprim-sulfamethoxazole 3%, ciprofloxacin 3%, and meropenem 2% were found resistant in that study, in Salmonella spp isolates (n = 326). Kesli et al. detected that all strains were susceptible to amikacin and cefazolin (36). When they sorted out the percentages of isolates, Salmonella serotype enteritidis 171 (52.4%) was found in the first place. Then, ranking continued as follows: Salmonella serotype typhimurium, 143 (43.9%), Salmonella serotype gallinarum, 7 (2.1%) and Salmonella serotype paratyphi A, 5 (1.5%). Erdem et al. reported an increase in resistance to ciprofloxacin (MIC > or = 0.125 mg/L) in S. typhimurium, S. paratyphi B, and S. enteritidis strains and that result has an emerging problem in S. enterica in Turkey (17).
As indicated by different studies around the world, an overall significant increase in resistance to ampicillin (14% to 50%), and nalidixic acid (10%), tetracycline (30%), and erythromycin (72%) was reported from different regions (37). Fortunately, trimethoprim/sulphamethoxazole, gentamicin, kanamycin, and chloramphenicol were reported susceptible in many areas. Chu et al. reported that S. enterica serotype enteritidis isolates (serotype 1a) has remained relatively more drug-susceptible than other common serotypes derived from human sources, such as S. enterica serotypes typhimurium (37). It was also reported that ciprofloxacin resistance rate was 8.05% (147/1826) in China (38). Resistance to ampicillin, chloramphenicol, and nalidixic acid were 20%, 11%, and 2.7%, respectively, in the United States (39). Surveillance programmes should be tracked carefully because of the extensive use of antibiotics in livestock production (40, 41).
6. Conclusion
In summary, it is recommended for health authorities to follow the regulations of the tracking food poisoning outbreaks. In food poisoning, the manual for infectious specimen collection must be known to collect samples from possible sources and food handlers in order to meet the need for disease prevention. This will help identify the true source of the disease. Restaurant and kitchen workers need to pay attention to personal and food hygiene, and to stop handling food when they have gastrointestinal symptoms. This is the best way to prevent food poisoning.