1. Background
Abortion is the unplanned and natural ending of a pregnancy occurring before the 20th week, usually involving a fetus weighing approximately 500 g. In the context of abortion, infectious agents have been shown to exert noteworthy efficacy (1, 2). Fatal pregnancy termination has a high prevalence in both developed and impoverished countries. This prevalence is reported as 3.9%, involving Group B Streptococcus (GBS) in women of childbearing age globally (3). Different statistics have been reported for abortion in Iran, but most of them are near global statistics (4). Congenital infections pose a significant threat to fetal health in humans, with bacterial agents emerging as a crucial contributor (5). Several bacterial agents have been identified as common causes of abortion. These include Mycoplasma hominis, M. genitalium, Ureaplasma urealyticum, Streptococcus agalactiae, Chlamydia trachomatis, Listeria monocytogenes, Brucella, and Helicobacter pylori (6-8).
A wide range of noteworthy events and advancements have emerged during the calendar year of 2021. Among the common bacterial factors involved in abortion, S. agalactiae (as a GBS member) is an important agent. Streptococcus agalactiae is aerobic, capsule-forming, Gram-positive cocci among the normal flora of the urinary-genital and lower digestive tract of many adults, so it has been isolated from the urinary-genital and digestive tracts of about 25% of healthy adult women (9). A meta-analysis determined that approximately 19.7 million pregnant women were affected by rectovaginal colonization with GBS in 2020 (5). The colonization of the bacterium in pregnant women generally remains without noticeable symptoms; however, it has the potential to lead to urinary tract infections, septicemia, cystitis, pyelonephritis, wound infections, chorioamnionitis, endometritis, miscarriages, and labor fever (10, 11). Recent research has demonstrated a gradual escalation of antibiotic resistance in numerous nations with respect to GBS strains (12-14).
The identification of bacterial infections in women who have experienced miscarriages and those within the childbearing age could potentially mitigate the financial burden on patients and the healthcare system stemming from the administration of antibiotic treatment for these infections (2, 7). Therefore, this study was designed to investigate the prevalence of three abundant infections in the vaginal secretions of women with abortion. On the other hand, GBS serotypes have a high presence in many geographic regions, and some serotypes are most prevalent in specific areas. Therefore, continuous monitoring studies, such as this study, are essential for determining the distribution of capsule serotypes in order to design specific antigen vaccination and treatment protocols optimally (15). In relation to the geographical distribution of S. agalactiae serotypes, it has been reported that serotype III is the most common type in Europe, Central Asia, Africa, and Australia. Other serotypes that have been reported in low-income countries were serotypes I, II, and V, in sequence (16, 17).
2. Objectives
This study was conducted in Isfahan, Iran, to determine the prevalence of M. hominis, L. monocytogenes, and S. agalactiae in the vaginal secretions of women who have had a history of miscarriage by multiplex PCR. Furthermore, the types of capsular antigen were detected in S. agalactiae isolates by multiplex PCR and sequencing. Additionally, it was sought to assess the impact of certain risk factors on miscarriage in these women, utilizing demographic information obtained from the patients.
3. Methods
3.1. Isolation of the Bacteria from Vaginal Samples
The bacterial samples studied in this study included 110 samples of vaginal secretions from women who had 2 - 3 previous abortions and were referred to gynecological departments because of a sudden abortion. A proven cause of abortions in these women had not been detected. Also, 110 samples were taken from pregnant women without reproductive problems or genital infections. The Cochran formula was used for the determination of the sample size, and a cluster sampling method was performed. The studied women were all married, aged between 21 and 35 years, with no assisted reproductive methods for pregnancy. These women had been referred to different medical centers with a gynecology department in Isfahan, Iran. The samples were taken immediately after abortion from the middle third area of the vagina with sterile swabs by gynecologists. They were transferred to the microbiology laboratory in 0.9% normal saline on ice for use in culturing and Todd Hewitt Broth medium (Merck, Germany) as a transfer medium (18). At the time of sampling, a questionnaire was completed to collect the demographic information of the patients, including education and economic status, age, blood groups, weight during pregnancy, the number of abortions, and the patient’s stress status and vitamin D level.
3.2. Designing the Multiplex Polymerase Chain Reaction Protocol for the Identification of the Isolates
All chemicals were obtained from Merck, Germany. All isolates were kept at - 70°C. The DNA was extracted from each isolated bacterium by phenol-chloroform method (19, 20). The multiplex Polymerase Chain Reaction (PCR) was used for the identification of the isolates based on the sequence of the 16S rRNA gene in the extracted DNA from each isolate by using specific primers. The sequences of the primers and their relevant reference strains are shown in Appendix 1. The PCRs were done by using the reaction mixture (CinaClon, Iran) containing 1X PCR buffer, 1.5 mM MgCl2, 200 µM dNTPs, 0.4 µM of each primer, 1-unit Taq DNA polymerase, and 0.5 µg/mL of the template DNA in a total volume of 25 µL in a T100 thermal cycler (Bio-Rad, USA).
The reaction protocols included an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 35 s, primers’ annealing at the temperatures and times obtained in gradient reactions for each primer, and extension at 72°C for 35 s. Each reaction was ended by a final extension at 72°C for 5 min. The PCR products were finally visualized by 1% agarose gel electrophoresis (NOGEN, Iran) and confirmed in polyacrylamide gel electrophoresis (NOGEN, Iran). The PCR products were sequenced (BIONEER, South Korea), and the obtained sequences were analyzed using the BLAST server to find the homologies in GenBank. The confirmed sequences were deposited in the NCBI genomic database. According to the Tm of the primers and the length of PCR products, a multiplex PCR protocol was designed for the simultaneous diagnosis of three infections by M. hominis, L. monocytogenes, and S. agalactiae. For this purpose, different concentrations of DNAs from reference strains were used as templates in multiple PCR to obtain the best amplification results.
3.3. Capsular Antigen Detection
The capsular polysaccharide type in S. agalactiae isolates was detected by a multiplex PCR method on the bacterial purified DNA using specific oligonucleotide primers presented in Appendix 2. The PCR temperature protocol included an initial step at 94°C for 3 min, followed by 40 repetitive cycles of 94°C for 45 s, 54°C for 40 s, and 72°C for 30 s each, and a final step at 72°C for 10 min. The evolutionary history was inferred by using the maximum likelihood method and JTT matrix-based model. All positions with less than 95% site coverage were eliminated. Evolutionary analyses were conducted in MEGA X.
3.4. Statistical Analysis
Descriptive statistics, including numbers, percentages, means, and standard deviation, and inferential (analytical) statistics, including chi-square, independent t, and Fisher’s exact tests, were employed. Also, SPSS 22 software was used for statistical analysis of data. The graphs were drawn using Excel software.
4. Results
4.1. Abortion Risk Factors in the Studied Women
The highest frequency of blood groups in the examined samples was related to blood group O+ in both groups, and the lowest frequency was related to blood group AB- in both groups, with no significant differences between blood groups (P = 0.05) (Appendix 3). The results of examining some other abortion risk factors in the studied women are shown in Appendix 4. Between healthy women who have had an abortion and even their husbands, the frequency of high school graduates and academic education was higher than the high school undergraduated women, while there was no significant difference between them. As the results showed, moderate economic status was most common in both aborted and healthy pregnant groups. There was no significant difference in the frequency distribution of women based on economic status between the healthy pregnant group and the group with abortions.
The results showed no significant difference between the two groups in terms of having a history of cesarean section. Also, no significant difference was seen between the two groups in terms of overweight. The stress during pregnancy was significantly greater in women with abortions than in healthy pregnant women. Concerning vitamin D levels, the results showed that the chance of miscarriage in women with normal vitamin D levels was significantly lower than that in women with vitamin D deficiency. The results of urinary infection, vaginal infection, and co-infection examination in the 2 groups of studied women are presented in Table 1. The women with abortions were significantly more infected than the healthy pregnant women. The women who had infections in both groups were sampled for isolation and identification of bacteria.
| Variables | No. (%) | Statistics | P-Value | Odds Ratio (95% Confidence Level) | |
|---|---|---|---|---|---|
| Healthy Pregnant | With Abortion | ||||
| Urinary infection | 29.66 (6.94 - 126.73) | ||||
| No | 108 (98.2) | 71 (64.5) | |||
| Yes | 2 (1.8) | 39 (35.5) | 41.038 | 0.001 | |
| Vaginal infection | 22.029 (6.57 - 73.88) | ||||
| No | 107 (97.3) | 68 (61.8) | |||
| Yes | 3 (2.7) | 42 (38.2) | 42.491 | 0.009 | |
| Double infection | 21.33 (2.79 - 163.82) | ||||
| No | 110 (100) | 92 (83.6) | |||
| Yes | 0.0 (0.0) | 18 (16.4) | 16.648 | 0.008 | |
| Total | 110 | 110 | 100 | 100 | |
Frequency Distribution of Various Types of Infections in Two Groups of Patients and Healthy Pregnant Women
4.2. Bacterial Isolates
The results are shown in Table 2. Based on the result of Fisher’s exact test, significant differences were seen between the two groups of abortion and healthy pregnant women in terms of the presence of L. monocytogenes and M. hominis. In comparison, there was no significant difference between the two groups in terms of the presence of S. agalactiae.
| Bacterial Isolate | No. (%) | Statistics | P-Value | Odds Ratio (95% Confidence Level) | |
|---|---|---|---|---|---|
| Healthy Pregnant | With Abortion | ||||
| Mycoplasma hominis | --- | 0.489 | --- | ||
| Negative | 110 (100) | 105 (95.46) | |||
| Positive | 0 (0.00) | 5 (4.54) | |||
| Listeria monocytogenes | --- | 0.622 | --- | ||
| Negative | 110 (100) | 107 (97.3) | |||
| Positive | 0 (0.00) | 3 (2.7) | |||
| Streptococcus agalactiae | 7.135 | 0.008 | 3.270 (1.8 - 321.091) | ||
| Negative | 109 (99.09) | 90 (81.81) | |||
| Positive | 1 (0.90) | 10 (9.09) | |||
Frequency Distribution of Abortion-causing Bacteria in Two Groups of Patients and Controls
4.3. Designed Multiplex PCR Protocol
A multiplex PCR was designed for the simultaneous amplification of the 16S rRNA gene in M. hominis (a 213 bp fragment), L. monocytogenes (a 417 bp fragment), and S. agalactiae (a 331 bp fragment). The results for the detection of the strains in some samples with different concentrations of template DNA are shown in Appendix 5.
4.4. Capsular Antigen Detection in Streptococcus agalactiae Isolates
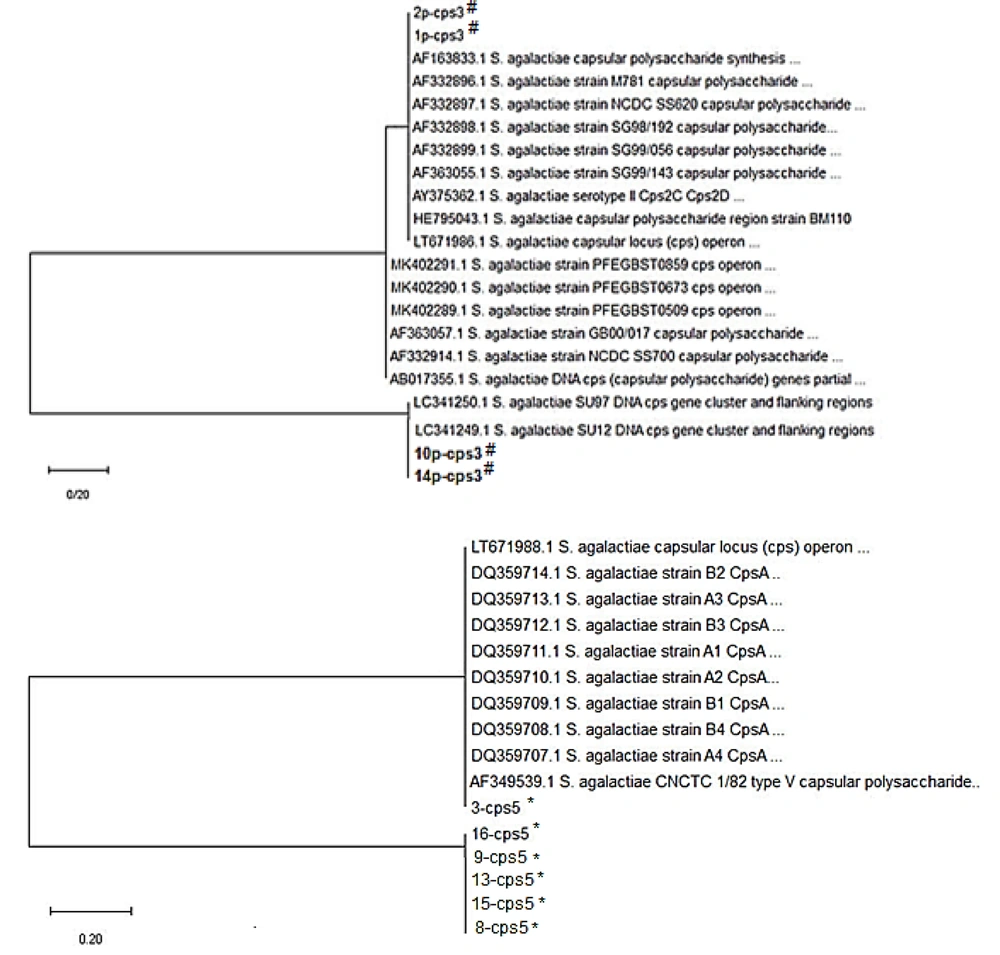
In a total of 10 isolates of S. agalactiae, the caps III and caps V genes were identified with frequencies of 40% (4 isolates) and 60% (6 isolates), respectively. The phylogenetic trees are shown in Figure 1. As shown, no difference was seen in the sequences of each of the caps III or caps V genes.
5. Discussion
The presence of genital infections has been associated with impaired fertility (21). Diagnosing bacterial infection in women can prevent more miscarriages and decrease its economic and health impacts on women in society (22). Several risk factors may predispose a pregnant woman to early pregnancy loss, among which infection is one of the most important factors (23). In this study, a novel multiplex PCR method was used for the simultaneous detection of 3 bacteria: M. hominis, L. monocytogenes, and S. agalactiae, which can be used as a complementary method that has been proposed previously for the simultaneous detection of other vaginal infections related to abortion (24-26). In the present study, S. agalactiae, M. hominis, and L. monocytogenes were detected in the vaginal secretions of women with abortion in the frequencies of 4.54%, 2.7%, and 9.09%, respectively. Mycoplasma hominis and L. monocytogenes were not found in the vaginal secretions of healthy pregnant women. The frequency of S. agalactiae in the vaginal secretions of those women was significantly lower than that in the women with abortion. In another study in Iran, the frequency of GBS was 7% in the samples taken from the vaginal secretions of 100 pregnant women (27).
It has been suggested that infection with M. hominis may be a common cause of spontaneous abortion (28). It has been reported that the colonization rate of these two bacteria in patients with miscarriage was higher than their frequencies in normal pregnancy groups. These bacteria are opportunistic pathogens of the urinary tract that are able to change the microenvironment of the uterine cavity against the normal growth of the fetus by causing multiple infections (29). In a study, the incidence of L. monocytogenes infection was found to be 3.66% among women with spontaneous abortion, 1.83% among women with normal delivery, 3% among fertile women, and 0% among infertile women (30). In another study, the prevalence of L. monocytogenes contributing to human spontaneous abortions was determined to be 14.8% (31). In another study, the correlation between M. hominis and spontaneous abortion in gravid women was assessed. The M. hominis infection (spontaneous abortion) was found in 2 individuals in the case group, constituting 1.83% of the group’s population.
In both the case and control groups, there was no observed correlation between M. hominis infection and spontaneous abortion (32). In the present study, the serotypes III (4%) and V (6%) were prevalent in the S. agalactiae isolates. This prevalence was similar to the prevalence that has been reported in Asia, although in some regions, serotypes I and II also have been reported (16, 17). The fact that the sequences of each caps III or caps V gene did not have any differences among the S. agalactiae isolates can be of concern for the health care system and vaccine designers for controlling the abortions caused by this bacterial infection.
5.1. Conclusions
The application of a previously developed novel multiplex PCR method is proposed for the concurrent identification of L. monocytogenes, M. hominis, and S. agalactiae in women with a history of abortion. In this study, the sequences of the caps genes had no difference, which is promising for adopting vaccination and therapeutic strategies.