1. Background
Klebsiella is a Gram-negative bacterium that is ubiquitous in various environments and belongs to the Enterobacteriaceae family (1). It is more likely to cause acquired infections in hospitalized patients and immunosuppressed individuals (2). Among the Klebsiella genus, K. pneumoniae is the most frequently identified species and is responsible for the majority of nosocomial infections (3). Klebsiella pneumoniae accounts for over 70% of human Klebsiella infections, leading to conditions such as pneumonia, bacteremia, sepsis, meningitis, and diarrhea (4). Additionally, K. pneumoniae is the second most common cause of urinary tract infections (UTIs) after Escherichia coli (5).
Beta-lactam antibiotics are the most commonly used antibacterial agents for treating infectious diseases. Approximately 65% of injectable antibiotics in the United States fall into this category (6). These antibiotics hinder the biosynthesis of peptidoglycan by inactivating penicillin-binding proteins (PBPs) (7). Among beta-lactams, carbapenems are highly utilized in hospitals due to their potent broad-spectrum antibacterial activity and minimal side effects (8). The global concern lies in the emergence and prevalence of carbapenem resistance (9).
Carbapenems are typically reserved as a last resort for treating severe infections caused by multidrug-resistant gram-negative bacilli (MDR-GNB). Resistance to carbapenems in Gram-negative bacteria primarily arises from the production of carbapenemase enzymes (10). These enzymes are categorized into three distinct groups, A, B, and D, based on Ambler's classification (11, 12). Currently, the most significant serine carbapenemase enzymes in class A are the K. pneumoniae carbapenemase (KPC) enzymes (13). Diagnostic methods for detecting KPC enzymes include both phenotypic and molecular approaches (7). In 2017, the Clinical & Laboratory Standards Institute (CLSI) recommended the use of the modified carbapenem inactivation method (mCIM), which boasts a sensitivity and specificity of over 99% for detecting carbapenemases among Enterobacteriaceae. mCIM has gained popularity in microbiological laboratories due to its simplicity, high sensitivity, specificity, widespread applicability, and cost-effectiveness (14).
2. Objectives
In our current research, our aim was to investigate the prevalence and expression levels of carbapenemase-encoding genes in carbapenem-resistant K. pneumoniae (Cr-KPN) strains isolated from patients admitted to teaching hospitals in Shiraz, Iran. Recognizing the significance of Cr-KPN in nosocomial and community-acquired infections, we assessed carbapenemase production and resistance using mCIM and minimum inhibitory concentration (MIC) tests, respectively. Additionally, we utilized multiplex-PCR and real-time PCR assays to determine the frequency and expression levels of carbapenemase-encoding genes.
3. Methods
3.1. Study Design and Identification of Bacteria
This study was a cross-sectional investigation conducted on all patients who had been hospitalized in the internal wards and intensive care units (ICUs) of two teaching hospitals, specifically Faghihi and Namazi hospitals, located in Shiraz, Iran. Sample collection took place over a six-month period, spanning from September 2021 to February 2021. All clinical samples referred from hospital wards to clinical laboratories were included in this study. Samples collected from patients who had taken antibiotics in the last week were considered, while samples taken from urine bags were excluded from the study.
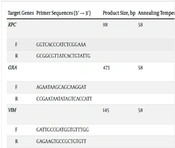
To initially identify K. pneumoniae, a series of conventional biochemical tests were conducted. These tests included Gram staining, the oxidase test, assessment of H2S and urease production, evaluation of motility, lactose fermentation, IMViC tests (Indole test, Methyl Red test, Voges Proskauer test, and Citrate utilization test), as well as nitrate reduction (15). Genomic DNA from the isolates was extracted using a DNA extraction kit (Tissue Genomic DNA Extraction Mini Kit, FAVORGEN/Taiwan). To confirm the final identification of the K. pneumoniae isolates, a polymerase chain reaction (PCR) assay targeting the specific rcsA gene was employed (16). The primer sequence used is provided in Table 1.
| Target Genes | Primer Sequences (5′ → 3′) | Product Size, bp | Annealing Temperature, °C | Reference |
|---|---|---|---|---|
| KPC | 88 | 58 | This study | |
| F | GGTCACCCATCTCGGAAA | |||
| R | GCGGCGTTATCACTGTATTG | |||
| OXA | 473 | 58 | This study | |
| F | AGAATAAGCAGCAAGGAT | |||
| R | CCGAATAATATAGTCACCATT | |||
| VIM | 145 | 58 | This study | |
| F | GATTGCCGATGGTGTTTGG | |||
| R | GAGAAGTGCCGCTGTGTT | |||
| IMP | 131 | 57 | This study | |
| F | TTGACACTCCATTTACTG | |||
| R | ATTAAGCCACTCTATTCC | |||
| IMI | 389 | 57 | This study | |
| F | GTAGAATAGCCATCTTGTT | |||
| R | ATTAGTAGCACCATTGTC | |||
| GIM | 231 | 58 | This study | |
| F | GTCAATTCCTACATACACATCAGA | |||
| R | CCTAAGCCTTCCCACTCA | |||
| NDM | 53 | 58 | This study | |
| F | GCCGACACTGAGCACTAC | |||
| R | GGGAACGCCGCACCAAAC | |||
| IMI-qPCR | 198 | 57 | This study | |
| F | TGAACGATTTCCATTATGTAGT | |||
| R | TATTGTAAAGCGGCAGCA | |||
| OXA-qPCR | 81 | 57 | This study | |
| F | TGCGTGTATTAGCCTTATCG | |||
| R | TTTCTTGCCATTCCTTTGC | |||
| 16SrRNA | 80 | 57 | This study | |
| F | GGGCTCAACCTGGGAACTG | |||
| R | TCACCGCTACACCTGGAAT | |||
| rcsA | 133 | 57 | This study | |
| F | GCTGTTTGTTATCTTTATGTC | |||
| R | AATCACCTGCTTATTATGC |
3.2. Antimicrobial Susceptibility Test
The determination of the MIC followed the broth microdilution method as outlined in the CLSI guideline (CLSI 2020) for both imipenem and meropenem (17). In brief, the MIC test was conducted using a range of doses from 64 to 0.125 µg/mL in U-shaped 96-well microplates, and quality control was maintained using E. coli ATCC 25922 (18).
3.3. Phenotypical Screening of Carbapenemase Production
The phenotypic identification of carbapenemase-producing K. pneumoniae was carried out through the mCIM assay, following the CLSI guidelines. To summarize, a 1 µL loopful of bacteria from an overnight blood agar plate was mixed in a tube containing 2 mL of TSB medium and vortexed for 10 - 15 seconds. Subsequently, a 10 µg meropenem disk (Liofilchem, Italy) was added to each tube using sterile forceps. The tubes were then incubated at 35°C in ambient air for 4 hours. After this incubation period, the disks were removed from the tubes, and excess liquid was expelled from the disk before placing them on MHA plates that had previously been inoculated with meropenem-susceptible E. coli ATCC® 25922 as an indicator strain. All plates were incubated at 35°C overnight. Results were interpreted as follows: Carbapenemase positive (only mCIM positive) if the zone diameter was 6 - 15 mm and carbapenemase negative (mCIM negative) if the zone size was ≥ 19 mm. Klebsiella pneumoniae ATCC BAA-1705 and K. pneumoniae ATCC BAA-1706 served as the positive and negative controls, respectively.
3.4. Detection of Carbapenemase-Encoding Genes
Genomic DNA extraction was carried out using a DNA extraction kit (Tissue Genomic DNA Extraction Mini Kit, FAVORGEN/Taiwan). The presence of carbapenemase-encoding genes, which include blaKPC, blaOXA-48, blaVIM, blaNDM, blaGIM, blaIMP, and blaIMI/NMC, was assessed through a multiplex PCR assay. The primer sequences utilized for detecting these carbapenemase-encoding genes can be found in Table 1. Amplification was conducted in a 25 μL reaction mixture comprising 12 μL of master mix (Amplicon, Pishgham, Iran), 1 μL of forward primer (10 pmol), 1 μL of reverse primer (10 pmol), 3 μL of template DNA, and 8 μL of sterile distilled water. The multiplex PCR reaction was carried out under the following conditions: Initial denaturation (one cycle at 95°C for 10 minutes), denaturation (94°C for 45 seconds), annealing (57 to 58°C for 45 seconds), extension (72°C for 60 seconds), and final extension (72°C for 7 minutes). Denaturation, annealing, and extension steps were repeated for 35 cycles. Positive PCR products were sent for sequencing, and the results were analyzed using Chromas software version 2.6.6 (http://www.technelysium.com.au) and the NCBI Nucleotide BLAST program available on the National Center for Biotechnology Information website (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
3.5. RNA Extraction and cDNA Synthesis
Total RNA was extracted from mCIM-positive isolates as well as carbapenem-resistant and intermediate isolates (imipenem and meropenem) using an RNA extraction kit (Total RNA Extraction Kit, Parstous Biotechnology Co./Mashhad/Iran). In brief, RNA was extracted based on the results obtained from mCIM and MIC testing from overnight cultures grown in a TSB medium containing sub-MIC concentrations of imipenem (1 µg/mL). RNA concentrations were measured using a Nanodrop device (Geneva Nano), and RNA concentrations were normalized using Diethyl pyrocarbonate water (DEPC water). Subsequently, cDNA synthesis was performed using a cDNA synthesis kit (Easy cDNA Synthesis Kit, Parstous Biotechnology Co., Mashhad/Iran).
3.6. Expression of Carbapenemase-Encoding Genes
The Real-time PCR method was employed to assess the expression levels of carbapenemase-encoding genes using specific primers (Table 1). These primers were designed using Allele ID 7.5 software and their specificity was confirmed through Primer Blast and Nucleotide Blast on the NCBI website. The study investigated how K. pneumoniae responded to environmental stress induced by sub-MIC levels of imipenem. Carbapenem-resistant isolates were divided into two groups: Group 1, which was exposed to 1 µg/mL of imipenem, and group 2, which was not induced by imipenem. Real-time PCR was conducted on a QuanStudioTM 3 system machine (Thermo Fisher, USA), and duplicate PCR reactions were carried out using RealQ Plus 2x Master Mix Green (Ampliqon, Denmark). The PCR conditions consisted of an initial activation step at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 20 seconds, annealing at the respective temperature for each gene for 25 seconds, and extension at 72°C for 30 seconds. Melt curve analysis was performed after 40 cycles to examine the Real-time PCR products. Data analysis was conducted using comparative Ct (∆∆Ct) values (19, 20).
3.7. Statistical Analysis
The results from MIC, mCIM, and ∆∆CT values were recorded in an SPSS file (Version 18.0, IBM SPSS Statistics for Windows, Armonk, NY, USA). Statistical analysis was carried out using the Chi-Square test, and P-values less than 0.05 were considered statistically significant (21).
4. Results
4.1. Patient Characteristics and Distribution of Samples
One hundred K. pneumoniae strains were isolated from a total of 671 different clinical samples collected from included patients, resulting in an overall frequency of K. pneumoniae strains of 14.9%. Among these strains, 58% were isolated from male samples, while 42% were from female samples. Regarding age groups, the highest prevalence of K. pneumoniae was observed in patients over 60 years old (n = 49/100; 49%), followed by those under 15 years old (n = 21/100; 21%), patients aged 31 to 60 years old (n = 13/100; 13%), and individuals aged 16-30 (n = 4/100; 4%).
The distribution of K. pneumoniae in various clinical samples was as follows: Urine (n = 44), blood (n = 19), sputum (n = 13), endotracheal tube (n = 12), abdominal fluid (n = 3), catheter (n = 3), abscess (n = 1), pulmonary (n = 1), axillary culture (n = 1), throat (n = 1), nasal discharge (n = 1), and wound (n = 1). Moreover, the characteristics and frequency of clinical specimens among different hospital wards are shown in Table 2. The highest number of urine samples (43.2%; n = 19/44) and endotracheal tube samples (58.4%; n = 7/12) were collected from the internal ward.
| Specimens | Hospital Wards | ||||||
|---|---|---|---|---|---|---|---|
| Internal | ICU | Infectious | Neurology | Hematology and oncology | Children | Surgery | |
| Urine (n = 44) | 19 (43.2) | 5 (11.4) | 13 (29.5) | 1 (2.3) | 1 (2.3) | 1 (2.3) | 4 (9) |
| Blood (n = 19) | 4 (21) | 5 (26.3) | 6 (31.6) | 0 (0.0) | 2 (10.5) | 1 (5.3) | 1 (5.3) |
| Sputum (n = 13) | 4 (30.7) | 5 (38.5) | 2 (15.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (15.4) |
| Endotracheal tube (n = 12 | 7 (58.4) | 3 (25) | 1 (8.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (8.3) |
| Abscess (n = 1) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Pulmonary (n = 1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100) |
| Central venous catheters (n = 3) | 0 (0.0) | 2 (66.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (33.3) |
| Axillary culture (n = 1) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Abdominal fluid (n = 3) | 0 (0.0) | 1 (33.33) | 1 (33.33) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (33.33) |
| Throat (n = 1) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Nasal discharge (n = 1) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Wound (n = 1) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Total (n = 100) | 36 (36) | 24 (24) | 23 (23) | 1 (1) | 3 (3) | 2 (2) | 11 (11) |
a Values are expressed as No. (%).
4.2. Meropenem and Imipenem MIC Results
Regarding meropenem and imipenem MIC results, 24% of K. pneumoniae isolates were resistant to imipenem, while 17% were resistant to meropenem (P = 0.319). Furthermore, results from the mCIM assay indicated that 26% of bacteria were positive for carbapenemase production, 6% were intermediate, and 68% were negative (P = 0.0001). Table 3 shows that there was no statistically significant relationship between mCIM results and hospital wards. Additionally, no statistically significant relationship was found between mCIM results and the type of samples (Table 3).
| Variables | mCIM Results | P-Value | ||
|---|---|---|---|---|
| Positive | Negative | Intermediate | ||
| Hospital Wards | 0.176 | |||
| Internal (n = 36) | 12 (33.3) | 24 (66.7) | 0 (0) | |
| ICU (n = 24) | 4 (16.7) | 17 (70.8) | 3 (12.5) | |
| Infectious (n = 23) | 7 (30.4) | 14 (60.9) | 2 (8.7) | |
| Children (n = 2) | 2 (100) | 0 (0) | 0 (0) | |
| Surgery (n = 11) | 0 (0) | 10 (90.9) | 1 (9.1) | |
| Neurology (n = 1) | 0 (0) | 1 (100) | 0 (0) | |
| Hematology and oncology (n = 3) | 1 (33.3) | 2 (66.7) | 0 (0) | |
| Total | 26 (26) | 68 (68) | 6 (6) | |
| Specimen type | 0.382 | |||
| Urine (n = 44) | 13 (29.5) | 31 (70.5) | 0 (0) | |
| Blood (n = 19) | 6 (31.6) | 11 (57.9) | 2 (10.5) | |
| Sputum (n = 13) | 1 (7.7) | 11 (84.6) | 1 (7.7) | |
| Endotracheal tube (n = 12) | 3 (25) | 6 (50) | 3 (25) | |
| Abscess (n = 1) | 0 (0) | 1 (100) | 0 (0) | |
| Pulmonary (n = 1) | 0 (0) | 1 (100) | 0 (0) | |
| CV catheters (n = 3) | 1 (33.3) | 2 (66.7) | 0 (0) | |
| Axillary culture (n = 1) | 0 (0) | 1 (100) | 0 (0) | |
| Abdominal fluid (n = 3) | 0 (0) | 3 (100) | 0 (0) | |
| Throat (n = 1) | 0 (0) | 1 (100) | 0 (0) | |
| Nasal discharge (n = 1) | 1 (100) | 0 (0) | 0 (0) | |
| Wound (n = 1) | 1 (100) | 0 (0) | 0 (0) | |
| Total (n = 100) | 26 (26) | 68 (68) | 6 (6) | |
a Values are expressed as No. (%).
4.3. Frequency of Carbapenemase-Encoding Genes
The presence of carbapenemase-encoding genes in K. pneumonia isolates was investigated, and the results showed that blaIMI/NMC, blaOXA-48, blaKPC, and blaNDM genes were detected in 91 (91%), 19 (19%), 11 (11%), and 2 (2%) of the isolates, respectively. However, blaVIM, blaGIM, and blaIMP genes were not found among the isolates. The distribution of these carbapenemase-encoding genes in different hospital wards is presented in Table 4. Among these genes, blaKPC had the highest frequency among K. pneumoniae isolates recovered from internal and ICU wards, accounting for 16.7% (n = 6/36) and 16.7% (n = 4/24), respectively. Additionally, 25% (n = 6/24) of K. pneumoniae isolated from the ICU ward were positive for the blaOXA-48 gene, while the blaNDM gene (5.6%; n = 2/36) was detected exclusively in the internal ward. Overall, there was no statistically significant relationship between the frequency of carbapenemase-encoding genes and hospital wards (P > 0.05).
| Hospital Wards | KPC | OXA-48 | NDM | IMI | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | P-Value | Positive | Negative | P-Value | Positive | Negative | P-Value | Positive | Negative | P-Value | |
| Internal (n = 36) | 6 (16.7) | 30 (83.3) | 0. 470 | 8 (22.2) | 28 (77.8) | 0.468 | 2 (5.6) | 34 (94.4) | 0.727 | 32 (88.9) | 4 (11.1) | 0. 941 |
| ICU (n = 24) | 4 (16.7) | 20 (83.3) | 6 (25) | 18 (75) | 0 (0) | 24 (100) | 21 (87.5) | 3 (12.5) | ||||
| Infectious (n = 23) | 0 (0) | 23 (100) | 4 (17.4) | 19 (82.6) | 0 (0) | 23 (100) | 22 (95.7) | 1 (4.3) | ||||
| Children (n = 2) | 0 (0) | 2 (100) | 1 (50) | 1 (50) | 0 (0) | 2 (100) | 2 (100) | 0 (0) | ||||
| Surgery (n = 11) | 1 (9.1) | 10 (90.9) | 0 (0) | 11 (100) | 0 (0) | 11 (100) | 10 (90.9) | 1 (9.1) | ||||
| Neurology (n = 1) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | ||||
| Hematology and oncology (n = 3) | 0 (0) | 3 (100) | 0 (0) | 3 (100) | 0 (0) | 3 (100) | 3 (100) | 0 (0) | ||||
| Total | 11 (11) | 89 (89) | 19 (19) | 81 (81) | 2 (2) | 98 (98) | 91 (91) | 9 (9) | ||||
a Values are expressed as No. (%).
Furthermore, 66.7% (n = 2/3) of K. pneumonia isolates obtained from CV catheter samples tested positive for both the blaKPC and blaOXA-48 genes. Additionally, 15.8% (n = 3/19) and 9.1% (n = 4/44) of K. pneumoniae isolated from blood and urine samples, respectively, were positive for the blaKPC gene. Conversely, 94.7% (n = 18/19) of K. pneumonia isolates from blood samples were positive for the blaIMI/NMC gene. There was no statistically significant relationship between the frequency of carbapenemase-encoding genes and the source of samples (P > 0.05). The analysis also revealed a statistically significant relationship between MIC results and the frequency of certain carbapenemase-encoding genes. As shown in Table 5, 62.5% (n = 15/24) and 8.3% (n = 2/24) of imipenem-resistant isolates were positive for the blaOXA-48 (P = 0.0001) and blaNDM (P = 0.040) genes, respectively. Conversely, 29.4% (n = 5/17), 76.5% (n = 13/17), and 11.8% (n = 2/17) of meropenem-resistant isolates were positive for blaKPC (p = 0.028), blaOXA-48 (P = 0.0001), and blaNDM (P = 0.007) genes, respectively.
| Susceptibility Profile (N) | KPC | P-Value | OXA-48 | P-Value | NDM | P-Value | IMI | P-Value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotics | P | N | P | N | P | N | P | N | ||||
| Imipenem | 0.199 | 0.0001 | 0.040 | 0.320 | ||||||||
| R (24) | 5 (20.8) | 19 (79.2) | 15 (62.5) | 9 (37.5) | 2 (8.33) | 22 (91.67) | 23 (95.8) | 1 (4.2) | ||||
| S (67) | 5 (7.5) | 62 (92.5) | 1 (1.5) | 66 (98.5) | 0 (0) | 67 (100) | 59 (88) | 8 (12) | ||||
| I (9) | 1 (11.1) | 8 (88.9) | 3 (33.33) | 6 (66.7) | 0 (0) | 9 (100) | 9 (100) | 0 (0) | ||||
| Meropenem | 0.028 | 0.001 | 0.007 | 0.355 | ||||||||
| R (17) | 5 (29.4) | 12 (70.6) | 13 (76.5) | 14 (23.5) | 2 (11.8) | 15 (88.2) | 17 (100) | 0 (0) | ||||
| S (72) | 5 (7) | 67 (93) | 1 (1.4) | 71 (98.6) | 0 (0) | 72 (100) | 64 (88.9) | 8 (11.1) | ||||
| I (11) | 1 (9) | 10 (91) | 5 (45.5) | 6 (54.5) | 0 (0) | 11 (100) | ||||||
Abbreviations: P, positive; N, negative; R, resistant; S, sensitive; I, intermediate.
a Values are expressed as No. (%).
4.4. Expression Levels of Carbapenemase-Encoding Genes
The results obtained from the real-time PCR assay indicated an increase in the expression levels of the blaKPC (1.66-fold), blaOXA-48 (7.30-fold), blaNDM (4.22-fold), and blaIMI/NMC (2.39-fold) genes in resistant isolates when exposed to imipenem. Our study demonstrated the overexpression of these genes when induced by 1 µg/ml of imipenem. It was observed that all bacteria resistant to imipenem in the MIC method expressed at least one of the investigated carbapenemase genes. Furthermore, resistant bacteria exhibited varying expression levels when exposed to sub-MIC values of imipenem, with isolates having higher MIC values generally showing higher expression levels. An increase in gene expression was noted in isolates that were identified as gene carriers by the PCR assay.
5. Discussion
Klebsiella pneumoniae is responsible for a significant proportion of nosocomial infections attributed to Klebsiella spp. Hospital-acquired pneumonia is associated with K. pneumoniae in 3.8% to 13.7% of cases (22). The primary cause of carbapenem resistance in Gram-negative bacteria is the production of carbapenemases (8). Among Gram-negative bacteria, Cr-KPN has emerged as a globally significant nosocomial pathogen (23). The objective of our investigation was to analyze the phenotype and molecular frequency of carbapenemase-encoding genes in Cr-KPN isolates from Shiraz, Iran. Additionally, Cr-KPN isolates underwent a real-time PCR assay to assess the expression levels of carbapenemase-encoding genes. Our analysis revealed a higher rate of resistance to imipenem compared to meropenem, suggesting that meropenem may be a more effective treatment option for Cr-KPN infections. The mCIM test indicated that 32% of isolates exhibited carbapenemase production with a positive or intermediate profile. The detection rates for the blaIMI/IMP, blaOXA-48, blaKPC, and blaNDM genes were 91%, 19%, 11%, and 2%, respectively.
Numerous studies worldwide have investigated the prevalence of carbapenemase-encoding genes in K. pneumoniae isolates. In 2023, Hallal Ferreira Raro et al. found that among K. pneumoniae producing carbapenemase, 40%, 28%, and 20% tested positive for the blaNDM-1, blaKPC, and blaOXA-48 genes, respectively (22). Solanki et al. reported that 71.1% of K. pneumoniae isolates were positive for the blaNDM-1 gene, while 14.3% were positive for blaKPC in a study conducted in the same year. Notably, all isolates tested negative for blaVIM and blaIMP genes (24). In 2022, Awoke et al. discovered that 29.6% of a total of 132 K. pneumoniae isolates were not susceptible to one or more carbapenems. Moreover, 21.2% of the isolates were found to produce carbapenemase, with 92.9% of these carrying the blaNDM gene (25). In a multinational prospective study, it was observed that 30% of 2301 K. pneumoniae samples produced carbapenemase (KPC, NDM, OXA-48-like, or VIM) (26).
Another study by Kilic et al. in 2016 reported that 50% of K. pneumoniae isolates were resistant to carbapenems. Additionally, 82.3%, 33.9%, 9.7%, and 4.8% of the isolates tested positive for the blaOXA-48, blaNDM-1, blaIMP-1, and blaVIM genes, respectively (27). Firoozeh et al. in 2016, similar to our findings, detected the blaKPC gene in 11.6% of K. pneumoniae isolates (28). In 2017, Khorvash et al. conducted a study on K. pneumoniae isolates, reporting a prevalence of 10.3% for blaVIM, 10.3% for blaIMP, and 3.4% for blaOXA genes. Notably, blaKPC and blaNDM genes were not detected (29). Hosseinzadeh et al. in 2018 revealed that among 29 carbapenem-resistant K. pneumoniae, 25 exhibited a high level of resistance to imipenem (MIC ≥ 4), with 10.9% and 0.9% of isolates testing positive for blaNDM-1 and blaOXA-48 genes, respectively (30).
Finally, in 2018, Moghadampour et al. conducted a study indicating that 91.9% of K. pneumoniae isolates were resistant to carbapenems. The frequencies of blaIMP, blaNDM-1, and blaOXA-48 genes were reported as 2.9%, 52.9%, and 70.6%, respectively (31). In 2020, Vivas et al. (32) found that 56.5% of K. pneumoniae isolates harbored at least one carbapenemase gene, with 50.3% carrying blaNDM-1 and 5.4% carrying blaKPC genes. Additionally, 1.2% of K. pneumoniae isolates possessed both blaNDM-1 and blaKPC genes. Their study employed the broth microdilution method to determine the MIC of K. pneumoniae isolates, revealing that isolates carrying blaNDM-1 and blaKPC genes exhibited high resistance to carbapenems.
In 2019, Aires -de-Sousa et al. conducted a study in Portugal (33), identifying blaKPC-3, blaOXA-181, and blaGES-5 as the most prevalent carbapenemase-encoding genes, with frequencies of 78%, 20%, and 17%, respectively. Among 46 carbapenemase-producing isolates, 28% were susceptible to imipenem (MIC < 0.5 mg/L), and 22% showed susceptibility to meropenem (MIC < 1 mg/L). In a 2020 study by Lopes et al. (34), blaKPC-2 (91%) and blaOXA-48 (9%) were identified as the most common carbapenemase-encoding genes. In our present study, real-time PCR analysis demonstrated that exposure to imipenem led to a 1.66-fold increase in the expression level of blaKPC, a 7.30-fold increase in blaOXA-48, a 4.22-fold increase in blaNDM, and a 2.39-fold increase in blaIMI/NMC genes. Notably, the expression levels of blaOXA-48 and blaNDM genes were higher than those of other genes. In a 2019 study by Mohammadi Bandari et al. (35), the expression levels of blaKPC and blaGES increased by 1.04-fold and 12.21-fold, respectively, in the presence of 2 µg/mL of imipenem. Their findings also indicated overexpression of these genes when induced by 2 µg/mL of imipenem, consistent with the results of our study.
5.1. Conclusions
Given the widespread resistance of K. pneumoniae strains to commonly used antibiotics, it is essential to detect isolates that produce carbapenemases to guide antibiotic treatment. The study conducted in the area revealed a worrisome prevalence of carbapenemase-producing K. pneumoniae isolates. These findings emphasize the significance of establishing active surveillance networks capable of monitoring and controlling the dissemination of carbapenemase-producing K. pneumoniae, which poses a global public health risk. Additionally, the data underscores the importance of implementing effective antimicrobial stewardship practices within hospitals.