1. Background
Antimicrobial research is crucial in addressing the global challenge of increasing resistance to β-lactam antibiotics, especially in urinary tract infections (UTIs) caused by gram-negative bacteria, particularly Escherichia coli. The rapid acquisition of antibiotic resistance and virulence genes in E. coli strains causing UTIs poses significant challenges for therapeutic interventions in healthcare systems (1, 2). The excessive and incorrect use of antibiotics has accelerated bacterial resistance evolution due to the selection pressure imposed (3). In 2019, a high prevalence of E. coli resistance to ciprofloxacin was reported in UTI isolates from Latin American patients (4), leading to an increase in the consumption of beta-lactam antimicrobials, such as cephalosporins, along with a corresponding rise in resistance rates.
This alarming trend prompted the World Health Organization (WHO) to classify extended-spectrum β-lactamase (ESBL)-producing E. coli as a priority pathogen for further research and surveillance (WHO, 2017) (5). Although, until the 1990s, the blaTEM and blaSHV genes were predominantly associated with beta-lactam resistance, since 2000, there has been a notable shift, with blaSHV being surpassed by blaCTX-M as the most frequently reported ESBL gene. The presence of both beta-lactamases is related to multidrug-resistant phenotypes, including resistance to beta-lactams, fluoroquinolones, and aminoglycosides (6, 7). The emergence of ESBL-producing E. coli strains, particularly those belonging to the O25b-ST131 clone, has further complicated the management of UTIs (1, 2). The prevalence of ESBL-producing E. coli in UTIs varies globally, influenced by several factors, including regional variations in antimicrobial usage practices (1, 8). In Mexico, there has been reported a high prevalence of ESBL-producing E. coli in clinical isolates and identified the O25b-ST131 clone, a pandemic clone associated with ESBL production (9-11).
The O25b-ST131 clone belongs to the B2 phylogenetic group and is known for its high level of antibiotic resistance and potential for causing recurrent UTIs (1, 12). Recurrent UTIs pose a substantial clinical challenge and contribute to the increased burden on healthcare systems (13). The high prevalence of ESBL-producing E. coli, particularly the O25b-ST131 clone, in urinary isolates from a Mexican healthcare institution underscores the importance of monitoring the molecular epidemiology, antibiotic resistance profiles, and transmission dynamics of these strains (11). These findings emphasize the regional variation in prevalence rates and highlight the urgent need to understand the molecular characteristics of the ESBL-producing E. coli O25b-ST131 clone. This study aimed to identify and molecularly characterize ESBL-producing E. coli isolates, with a specific focus on the pandemic O25b-ST131 clone associated with UTI in a healthcare institution in Mexico.
2. Objectives
The goal was to contribute valuable insights into the molecular epidemiology, antibiotic resistance profiles, and transmission dynamics of these strains, addressing the urgent need for understanding the regional variation in prevalence rates.
3. Methods
3.1. Bacterial Isolates and Antimicrobial Susceptibility Testing
A total of 254 E. coli isolates were obtained from a private hospital within October 2021 to June 2022 in Torreon, Coahuila Mexico. Bacterial species identification and antibiotic susceptibility tests were performed using the Vitek 2® (GN ID Card, BioMerieux, Francia). Isolates were identified as ESBL-producing strains by the double-disk synergism method according to the Clinical and Laboratory Standards Institute breakpoints (14).
3.2. Polymerase Chain Reaction Amplification Genes
Deoxyribonucleic acid (DNA) was obtained from colonies via heat shock. The ESBL-producing E. coli isolates were cultivated on lysogeny broth (LB) agar at 37°C overnight. The cells were subjected to two rounds of heat shock at 90°C for 10 minutes, followed by cooling at -20°C for 10 minutes. Subsequently, DNA was separated from cellular debris through centrifugation, and the resulting supernatant was collected for further experiments. The blaTEM, blaCTX-M-15, blaCTX-M-1 group, blaCTX-M-9 group, blaSHV, and blaGES genes were screened using specific primers for each gene by polymerase chain reaction (PCR) (Appendix 1) (15-17). The PCR conditions were conducted with an initial denaturation at 92°C for 2 minutes, followed by 30 cycles of denaturation at 92°C for 30 seconds, annealing at 58°C for 30 seconds, extension at 72°C for 45 seconds, and a final extension at 72°C for 5 minutes.
3.3. Escherichia coli Genotyping
To differentiate between commensal and pathogenic ESBL-producing E. coli isolates, phylogenetic group analysis was performed for each isolate by PCR as described by Clermont et al. The amplification of the chuA and yjaA genes, along with the tspE4C2 fragment, allowed the classification of the isolates as commensal or virulent extra-intestinal strains (18).
3.4. Genetic Characterization
Random amplified polymorphic DNA (RAPD) analysis was performed to identify the genetic diversity of qepA-positiveE. coli isolates. The RAPD was performed using decameric primers P1254 and PCR conditions described by Betancor et al. (19). The correlation by similarity was computed based on the band positions using the Dice coefficient, and the dendrogram was generated using the UPGMA clustering method, both within the GelCompar II program (20).
3.5. O25b-ST131 Identification
The ESBL-producing E. coli isolates were screened using primers O25pabBspe.F and O25pabBspe.R to amplify a 347 bp fragment of the pabB gene to identify isolates belonging to the O25b-ST131 clone. The identification of O25b typing was performed by PCR, as previously described by Clermont et al., using the rfbO25b.r and rfb1bis.f primers (21, 22).
4. Results
In the study conducted between 2021 and 2022 at the Torreon hospital in Mexico, 33.8% (86/254) of clinical isolates were identified as ESBL-producing E. coli. The confirmation of ESBL production was achieved through the double-disk synergism method. Among these isolates, 65.1% (56/86) and 34.8% (30/86) were from women and men, respectively. Approximately 68.6% (69/89) of the isolates were obtained from individuals, both male and female, predominantly aged over 60 years. Among the ESBL-producing E. coli isolates, 47% (41/86) were collected from nosocomial infections, and 52% (45/86) were from community-acquired infections. The primary sources identified in these isolates were UTIs at 75% (63/86) and blood infections at 10% (9/86). Women were the predominant demographic for UTIs, representing 79.4% (50/63) of these cases; however, men represented the remaining 20.6% (13/63).
The 63 ESBL-producing E. coli isolates from UTIs showed a resistant profile, with 100% of the isolates being resistant to ampicillin and cephalothin. Additionally, 96% (61/63) showed resistance to cefuroxime, cefotaxime, ceftazidime, ceftriaxone, and cefepime. Resistance to ciprofloxacin, norfloxacin and gentamicin was observed in 93% (59/63), 85% (54/63), and 39% (25/63) of the cases, respectively. Notably, no isolates from UTIs demonstrated resistance to amikacin, ertapenem, or meropenem.
Among the 86 ESBL-producing E. coli isolates, the main β-lactamase identified was blaCTX-M in 66% (57/86), followed by blaTEM in 8.1% (7/86), blaCTX-M/SHV in 5.8% (5/86), blaCTX-M/TEM in 4.6% (4/86), and blaSHV in 2.3% (2/86) (Table 1). The blaCTX-M, blaCTX-M/SHV, and blaCTX-M/TEM genotype were observed in 76.7% (66/86) of the ESBL-producing E. coli isolates, with a resistant profile of 100% (66/66) to ampicillin and cephalothin, 95.4% (63/66) to cefuroxime, cefotaxime, ceftazidime, ceftriaxone, and cefepime, 93.9% (62/66) to ciprofloxacin, 83.3% (55/66) to norfloxacin, and 40.9% (27/66) to gentamicin. Interestingly, the blaTEM and blaSHV genotypes were present in 10.4% (9/86) of the isolates, and all of them exhibited 100% (9/9) resistance to ampicillin, cephalothin, cefuroxime, cefotaxime, ceftazidime, ceftriaxone, cefepime, ciprofloxacin, and norfloxacin.
| Variables | n | Community-Acquired Infections (45) | Nosocomial Infections (41) | ||||
|---|---|---|---|---|---|---|---|
| Phylogenetic Group | β-lactamase | n | Phylogenetic Group | β-lactamase | n | ||
| Respiratory secretion | 3 | B2 | CTX-M-15 | 3 | |||
| Wound secretion | 6 | B2 | CTX-M-15 | 1 | B2 | CTX-M-15 | 1 |
| TEM | 1 | ||||||
| CTX-M-15/TEM | 1 | ND | 1 | ||||
| A | CTX-M-15 | 1 | |||||
| stool | 3 | B2 | CTX-M Group 9 | 1 | A | CTX-M-15 | 2 |
| UTI | 63 | B2 | CTX-M-15 | 13 | B2 | CTX-M-15 | 9 |
| TEM | 2 | TEM | 1 | ||||
| SHV | 2 | ND | 1 | ||||
| CTX-M-15/SHV | 2 | ||||||
| CTX-M-15/TEM | 1 | ||||||
| ND | 2 | ||||||
| D | CTX-M-15 | 3 | D | CTX-M-15 | 1 | ||
| ND | 1 | ||||||
| B1 | CTX-M-15 | 4 | B1 | CTX-M-15 | 2 | ||
| CTX-M Group 9 | 1 | CTX-M Group 1 | 1 | ||||
| CTX-M-15/SHV | 1 | ||||||
| CTX-M-15/TEM | 1 | ||||||
| A | CTX-M-15 | 7 | A | CTX-M-15 | 4 | ||
| TEM | 2 | ||||||
| SHV | 1 | ||||||
| CTX-M-15/SHV | 1 | ||||||
| ND | 1 | ||||||
| Blood | 9 | B2 | CTX-M-15 | 1 | |||
| CTX-M Group 9 | 1 | ||||||
| TEM | 1 | ||||||
| CTX-M-15/TEM | 1 | ||||||
| ND | 1 | ||||||
| D | CTX-M-15 | 1 | |||||
| A | CTX-M-15 | 2 | |||||
| TEM | 1 | ||||||
| Other samples | 2 | D | CTX-M-15 | 1 | |||
| A | CTX-M-15 | 1 | |||||
Characterization of ESBL-Producing E. coli Isolates
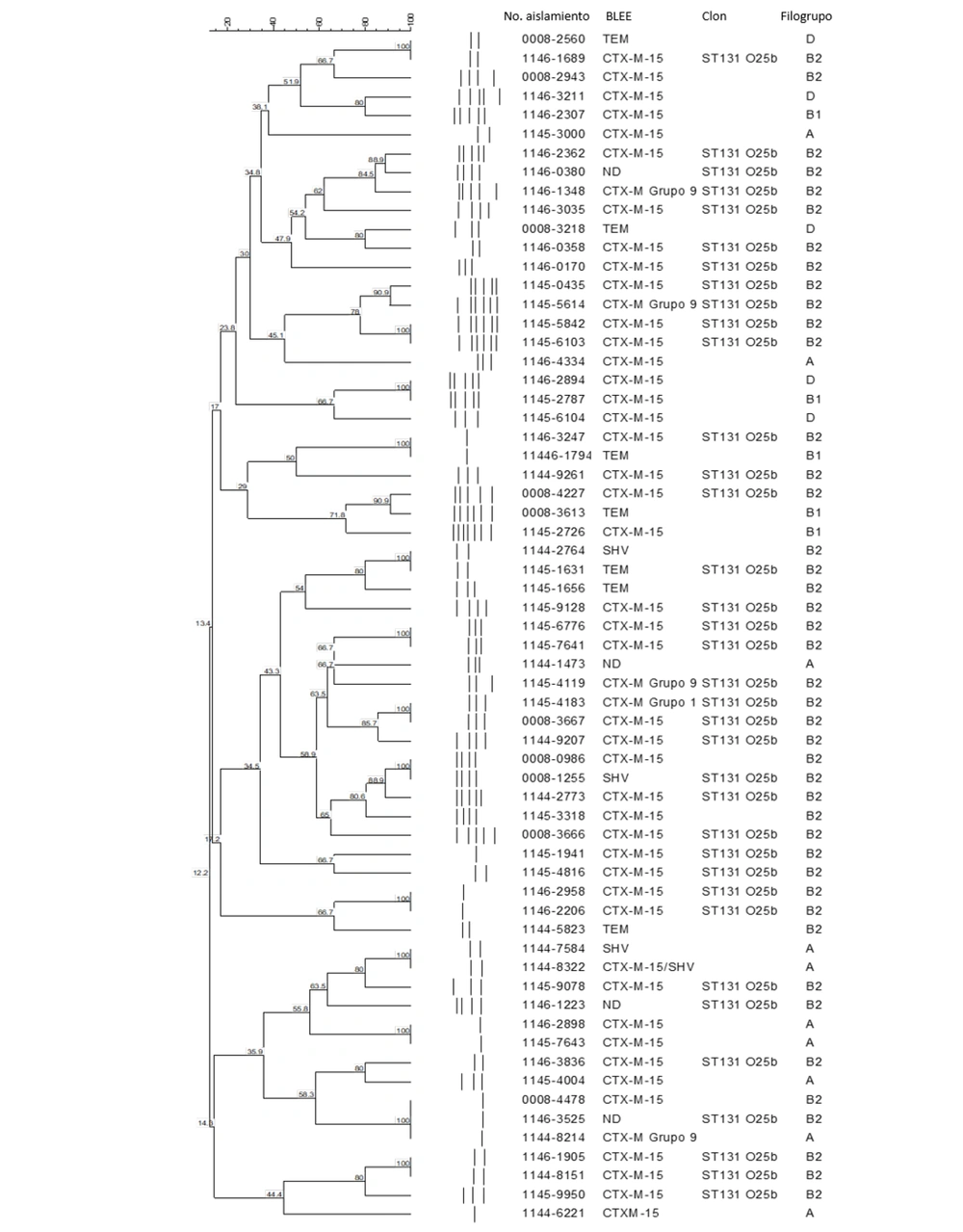
The ESBL-producing E. coli isolates were analyzed to identify the main phylogroups. The B2 phylogroup was the most prominent, comprising 54.4% (47/86) of the isolates, followed by group A with 25.5% (22/86), group B1 with 11.6% (10/86), and group D with 8.1% (7/86). Among the main phylogroups identified, B2 72.3% (34/47) and A 68.1% (15/22) were predominantly obtained from UTIs. Special attention was given to the main B2 phylogroup to identify the ESBL-producing E. coli isolates belonging to the O25b serotype and ST131. In the ESBL-producing E. coli isolates belonging to the B2 phylogroup, 40/86 (46.5%) were identified as the globally pandemic O25b-ST131 clone. However, in the analysis of genetic diversity and relationships among ESBL-producing E. coli isolates, no evident relationship was observed using RAPD between isolates with nosocomial infections and community-acquired infections (Figure 1).
5. Discussion
In recent years, E. coli has emerged as the primary pathogenic agent implicated in UTIs, becoming a critical challenge to antimicrobial research. Its rapid acquisition of antibiotic resistance and virulence genes, compounded by subsequent mutations, provides, enhanced its colonization abilities, complicated therapeutic interventions, and encouraged the WHO to designate E. coli as a pathogen of critical research priority (23). Urinary tract infections caused by gram-negative bacteria, especially E. coli, present a significant challenge globally. Recent studies have shed light on the resistance patterns of E. coli strains, which are the leading cause of UTIs.
In comparison to recent findings by the INVIFAR surveillance group, the present study highlights notable trends in antibiotic resistance, particularly focusing on ampicillin and cephalosporins. According to the INVIFAR report, there were high resistance rates: 80% to ampicillin and 30% to cephalosporins (24). The current study, in comparison, reveals resistance rates of 75% and 47%, respectively. Within the specific context of the Hermosillo healthcare system in Sonora, Mexico, the present study identified ESBL-producing Enterobacteriaceae. Notably, research conducted in four hospitals in this region showed that 11.6% of Enterobacteriaceae were ESBL producers, predominantly E. coli (95.6%) and Klebsiella pneumoniae (4.3%) being the predominant strains (25). These isolates exhibited alarmingly high resistance rates of 98% to cephalosporins, aztreonam, and ampicillin. Of particular significance is the substantial proportion (78%) of ESBL-producing isolates found in UTI samples obtained in the context of community-acquired infections (25). These findings are consistent with those of a previous study in Reynosa, Tamaulipas, and the current study, where 70% and 75% of the isolates were associated with UTIs, respectively (11). The consistent patterns observed in these studies correlate with the widespread presence of ESBL-producing E. coli found in UTIs, indicating a growing concern for antibiotic resistance.
The analysis of ESBL-producing E. coli in the present study revealed that 80% (70/86) of the isolates harbored the blaCTX-M gene, either alone or in combination with TEM, SHV, CTX-M group 1, and 9. Nevertheless, the presence of blaSHV and blaTEM alone was 10.4% (9/86) of the isolates. Similar results were previously reported in Mexico, with frequencies around 86-97% of producing blaCTX-M alone or with the co-production of blaSHV and/or blaTEM (6, 25, 26). The aforementioned findings demonstrate that the most prevalent ESBL genes in Mexico have undergone evolutionary changes over time, with frequencies shifting from the SHV and TEM gene families to the CTX-M gene family. Similar findings have been observed in a multicentric study conducted in Mexico over an eight-year period (6).
The emergence of E. coli as the primary pathogenic agent implicated in UTIs will be associated with the changes in the frequency of specific phylogroups, particularly B2. Notably, isolates belonging to the B2 phylogroup are frequently associated with extraintestinal infections, including UTIs. The frequency increase might correlate with the presence of more virulence-associated genes than the other phylogroups. This finding will be the main reason why 54% of the ESBL E. coli isolates in the current study belonged to the phylogenetic group B2. Notably, all isolates tested positive for O25b-ST131, a sequence type associated with a pandemic clone belonging to a B2 phylogroup. Similar observations were reported in a study on recurrent UTIs identified between 2004 - 2014 in a hospital in Sweden, where the phylogroup B2 was identified in 56% of the isolates, of which 78% belonged to ST131-O25b clone (8).
Previous studies have documented the frequency variations of ESBL E. coli isolates from UTIs that are associated with the B2 phylogroup and the global pandemic O25b-ST131 clone in Mexico. For instance, a study conducted in 2018 examined UTI isolates collected from two hospitals located in Tamaulipas, Mexico. The analysis revealed that the B2 phylogroup was present in 42% of the ESBL-producing E. coli isolates, with 23% of these isolates observed to be associated with the global pandemic O25b-ST131 clone (11). In the study conducted at the ISSSTE clinic in Guerrero, Mexico, in 2014, it was observed that 43% of the ESBL-producing E. coli isolates obtained from UTI samples belonged to the phylogroup B2. Furthermore, 37% of the isolates were observed to be the globally prevalent O25b-ST131 clone (27).
The current study demonstrated that the ESBL-producing E. coli isolates showed significant diversity in their clonal relationship patterns, forming around 35 groups. Of the 86 isolates, 73% showed a weak relationship; however, the rest of the isolates were not related, indicating the absence of specific outbreaks due to the presence of multiple groups. The aforementioned findings are comparable to those in Iran, where 14 groups were identified in the clonal relationship analysis of uropathogenic E. coli (28). Similarly, a study in Romania identified 13 groups with a 75% similarity; nevertheless, Ecuador exhibited extensive genetic diversity among these strains, resulting in more than 30 groups (29, 30).
The above-mentioned findings are similar to the findings of previous studies conducted globally, highlighting the widespread dissemination of this E. coli O25b-ST131 clone across Mexico and with similar distribution worldwide. Importantly, the present study provides valuable insights into the resistance landscape in Mexico. These observations call for collaborative efforts to combat the dissemination of these highly resistant clones. To better understand the local epidemiology, the findings of the present study are compared to the findings of studies conducted in various regions of Mexico. Consistently, there was a predominance of phylogenetic group B2 among E. coli strains recovered from UTI. Furthermore, the current investigation revealed significant clonal diversity among ESBL-producing E. coli strains. The formation of several distinct clonal groups highlights the intricate nature of their genetic relatedness. Interestingly, this diversity contributed to the absence of outbreaks, as multiple groups were present simultaneously. The aforementioned findings support the notion that the North region of Mexico harbors a diverse reservoir of ESBL-producing E. coli strains, necessitating targeted surveillance and control measures.
5.1. Conclusions
The present study revealed a concerning surge in antibiotic resistance among E. coli isolates, especially in UTIs, posing significant therapeutic challenges. Despite the observed increase in antibiotic resistance genes and enhanced colonization capabilities, definitive correlation with B2 phylogroup variations in E. coli's emergence as the predominant UTI pathogen requires further investigation. In conclusion, the urgent implementation of a comprehensive antimicrobial stewardship program in Mexico is crucial to address rising resistance rates, particularly among ESBL-producing E. coli strains. The identification of CTX-M-15-producing E. coli O25b-ST131 strains underscores the severity of this public health challenge. Collaborative efforts on national and international levels are essential to curb the spread of these highly resistant strains, emphasizing the rational use of antimicrobials in hospitals and communities to combat this pressing issue effectively.