1. Background
Cytolysin family is a single and highly homologous family and consists of species of 5 genera: Streptococcus, Bacillus, Clostridium, Listeria, and Arcanobacterium (1-7). Streptolysin O (SLO) from Streptococcus pyogenes is an oxygen-labile cholesterol-dependent cytolysin (2) and can oligomerize to form large pores (8-10) and cause the cytolysis of some eukaryotic cells, particularly erythrocytes (5) to bind to a cholesterol-containing membrane in the form of monomers. The produced pores allow the permeation of ions and small metabolites as well as macromolecules such as hemoglobin and immunoglobulin, with up to 100 kDa mass, to the cytosol (11).
The action for introducing proteins and nucleic acid into living cells has been found to have multifaceted applications in animal cell biology in humans or animals. With SLO, 3 active domains of large clostridial toxins (Clostridium difficile B-toxin, C. sordelli lethal toxin, and C. botulinum C2 toxin) were introduced into 3 different cell lines, and the results revealed that all the 3 toxins disrupted the actin cytoskeleton to cause the rounding up of the cells (7). A past study indicated that SLO treatment improved the cell fusion of fetal fibroblasts and the nucleic transferring of porcine embryo (12). Streptolysin O was used as an adjunct therapy with antibiotics to reduce the duration and severity of disease caused by Rhodococcus equipneumonia compared to antibiotic therapy alone (13). Adjunct therapy of SLO/antibiotics also reportedly modulated the site-specific expression of extracellular matrix and inflammatory genes in lungs of R. equi infected foals (14).
Most commercially available SLO was natural SLO purified from S. pyogenes cultures (15). However, the purification of natural SLO has several drawbacks such as low yield, considerable batch-to-batch variation, and biological risk in industrial processes. Therefore, recombinant SLO is used as an alternative to avoid most of those drawbacks. A great deal of research has been conducted to express full SLO protein, full mature SLO protein, and mature SLO fragment deficient in the N-terminal side.
The effects of different fusion tags of SLO expression in Escherichia coli was researched as follow. The recombinant mature SLO (containing 32 - 571 amino acids) fused with His-tag was expressed in E. coli, and the yield of the SLO fusion protein was 10 - 30 mg/L culture (16). Velazquez et al. expressed the fusion protein of SLO (78 - 571 acid amino sequence) with GST in E. coli, and its purified SLO yield was 1.5 mg/L culture (17). The recombinant mature SLO was expressed as another fusion protein with a thioredoxin and a C-terminal hexahistidine tag, and the yield of SLO fusion protein yield was 124 mg/L culture medium according to SLO activity concentration (2000 HU/mL cell suspension) and specific activity (16102 U/mg) (18). To meet the increasing need of SLO in research and application in animal cell biology and medicine, a PEL-SLO fusion protein was highly expressed with a pectate lyase (PEL) as fusion tag (19), and its hemolysis activity was characterized.
2. Objectives
A fusion protein of pectate lyase and streptolysin O were highly expressed with a pectate lyase (PEL, from Aspergillus nidulans) as a fusion partner. The conditions of expression, purification, and hemolysis characteristics of this fusion protein were researched.
3. Methods
3.1. Ethics Statement
The main materials of the research included bacteria, sheep red blood cell, and bacteria plasmids. The project was reported to Yancheng Teachers University Ethics Committee and showed that no animals, plants, and population were inclusive in the study (Yancheng Teachers University ethics committee code: reference number 2014030503).
3.2. Strain and Plasmids
The streptolysin O gene (slo, NCBI GenBank accession number: NC-011375) was from S. pyogenes NZ131. The pectate lyase gene (pel, NCBI GenBank accession number: EF452421) was from A. nidulans GR5 (Fungal Genetics Stock Center, Kanasas, MO, USA) (16). Escherichia coli strain DH5α (Lucigen, USA) was used as the clone host, and pMD18-T plasmid (TaKaRa, P.R. China) was employed as clone vector. Moreover, E. coli BL21 (DE3) (NEB, USA) was served as expression host and pET-28a (+) plasmid (Invitrogen, USA) was employed as expression vector. Sheep red blood cell (CELLBIO, China) was used to measure the hemolysis activity.
3.3. Designation of Amino Acid Sequence of Fusion Protein in Pectate Lyase and Streptolysin O
The amino acid sequence in fusion protein of pectate lyase and streptolysin O (PEL-SLO) is demonstrated in Figure 1. The amino acids 1 - 313 were a pectate lyase (PEL) from the mycelium of A. nidulans (19).
3.4. Cloning and Expression
The slo fragment was amplified with PfuUltraTM High-Fidelity DNA Polymerase (Stratagene, La Jolla, CA, USA), total genomic DNA from S. pyogenes NZ131, and sequence specific primers p1: ’-CATATGGCTTGCTCCCAAAGAAATGC-3’ 2: 5’-GGATCCCTACTTATAAGTAATCGAACC-3’) (Sangon Biontech, China) to construct slo/pMD18-T clone plasmid. The pel-ek-his DNA fragment was amplified with PfuUltraTM High-Fidelity DNA polymerase, pel/PMD-18T (19), slo/pMD18-T and sequence specific primers p3: 5’-CCATGGGCTCACCTGCGCCGGACCTCAACG-3’ p4: 5’-CATATG- GGTGATGATGATGATGATGATGATGATGATGC- TTGTCGTCGTCGTCCAATTTCT- GTCCGGCTGTACCCGGAATCACG-3’) (Sangon Biontech, China) to construct pel-ek-his/pMD18-T clone plasmid. Then, pel-ek-his-slo/pET-28a (+) was constructed and transformed into E. coli BL21 (DE3).
In Luria–Bertani (LB) with 1% (w/v) glucose and 50 mg/mL kanamycin, the transformants were cultivated at 37°C and 200 rpm to an absorbance (at 600 nm) of 0.8. Then, culture broth was added with Isopropyl ß-D-1-thiogalactopyranoside (IPTG) at 0.6 mM final concentration. The mixture was grown at 25°C and 200 rpm for another 36 hours for induction of recombinant protein synthesis.
3.5. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 11.0% polyacrylamide gel (20).
3.6. Extraction and Purification
Cell intracellular protein was extracted and loaded on a Ni2+-nitrilotriacetate-agarose column (Novagen, USA) to purify recombinant expressed PEL-SLO fusion protein according to the system manual. Eluted PEL-SLO was dialyzed against 10 mM phosphate buffer (pH 7.0), with a dialysis membrane (Sigma-Aldrich, USA). The amount of protein was measured using Bradford’s method (21).
3.7. Hemolysis Activity Assay
Hemolysis activity of PEL-SLO was measured as described by Herbert (22, 23).
4. Results
4.1. Expression
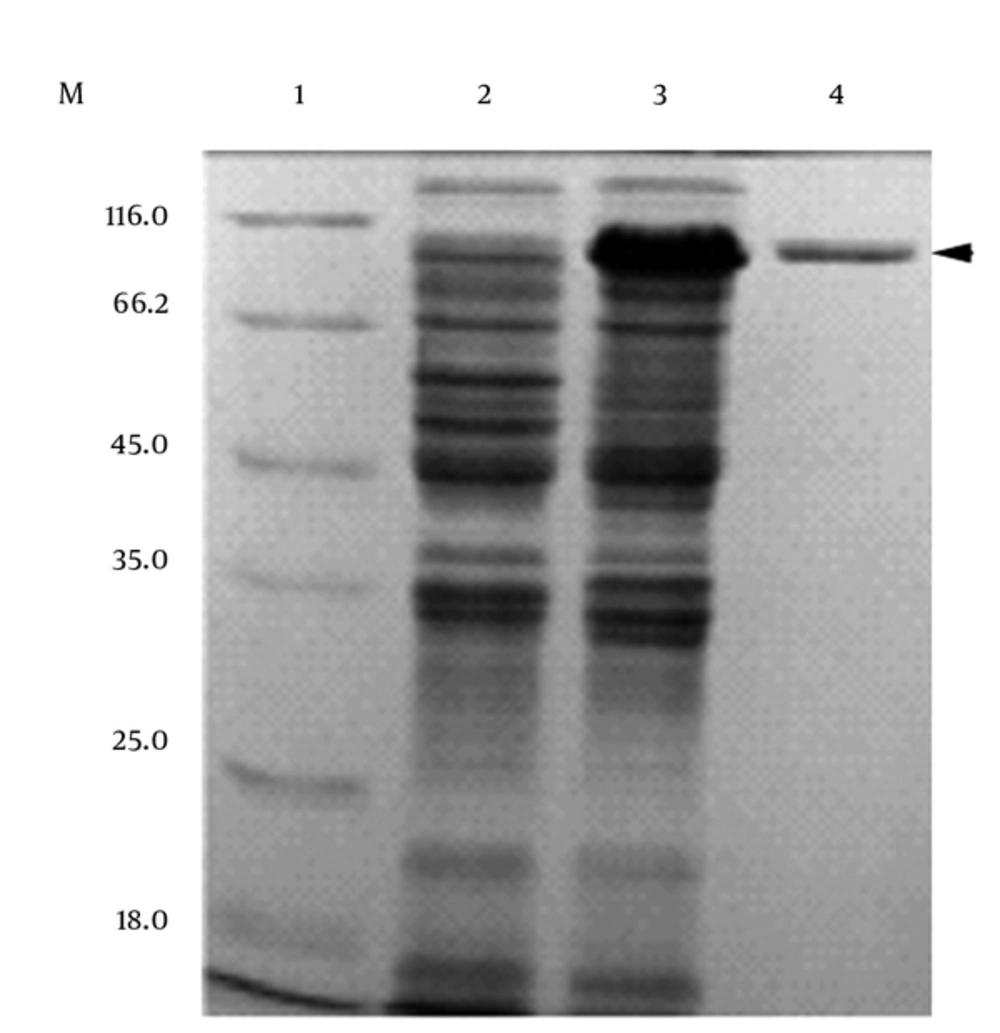
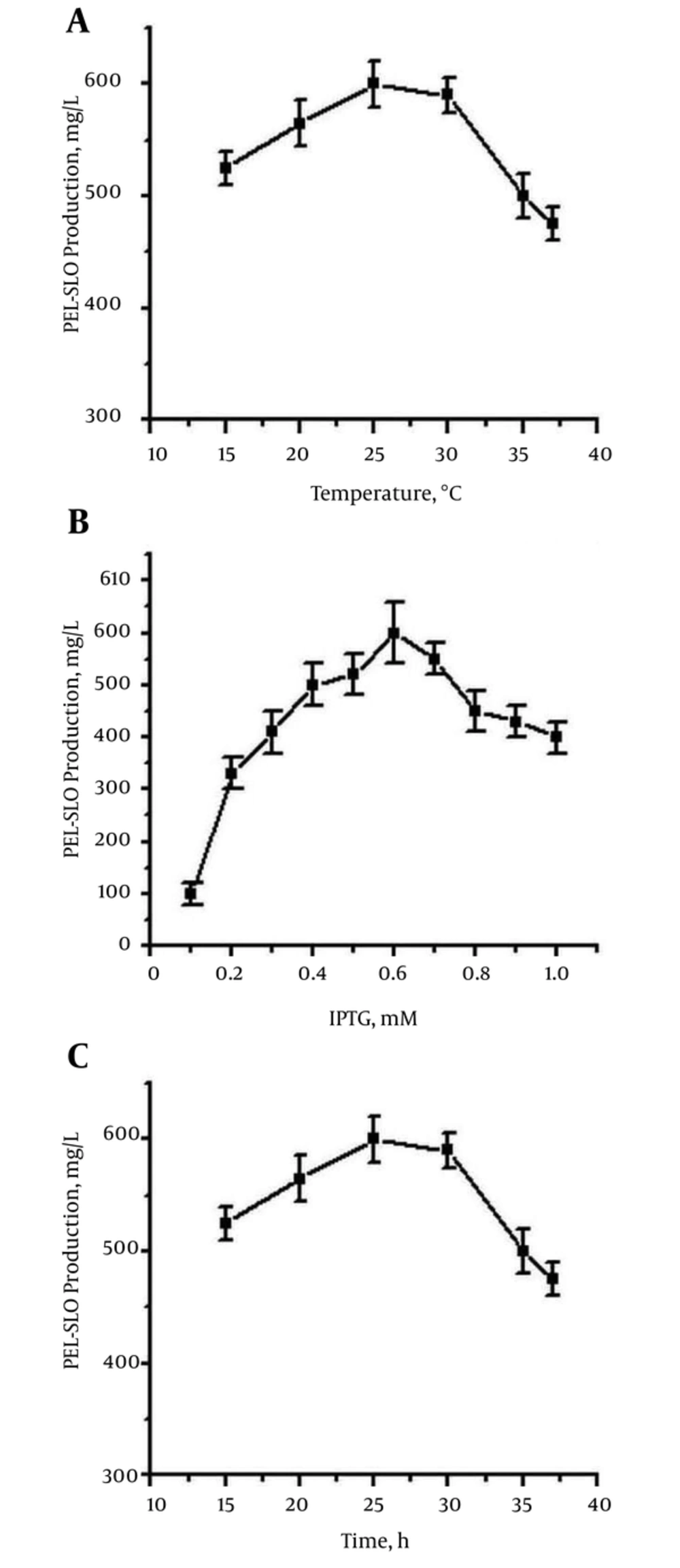
The recombinant PEL-SLO fusion protein was successfully overproduced (Figure 2). The recombinant SLO protein was also produced with E. coli BL21 (DE3) as the host for expression, and the pET-28a (+) plasmid as expression vector (Figure 2). After fully exploring the expression conditions, the best expression efficiency of soluble PEL-SLO fusion protein was found to be higher than 600 mg/L in shaking flask at 0.6 mM IPTG (Figure 3A), 25°C (Figure 3B), and for 36 hours (Figure 3C). The best expression efficiency of recombinant SLO (not using PEL as fusion partner) might be no more than 30 mg/L in shaking flask at 0.6 mM IPTG, 25°C, and for 36 hours (Figure 2).
M: Protein marker; 1: supernatant sample from induced pET-28a (+)/BL21(DE3); 2: supernatant sample from induced pel-ek-his-slo/BL21(DE3); 3: purified the fusion protein of pectate lyase and Streptolysin O by Ni2+-nitrilotriacetate-agarose column (loading buffer with 10 mmol/L imidazole binding; binding buffer: with 10 mmol/L imidazole; washing buffer: with 60 mmol/L imidazole; eluting buffer: with 100 - 500 mmol/L imidazole).
Cells of pel-ek-his-slo/pET-28a (+)/E. coli BL21 (DE3) were incubated at 37°C at 200 rev/min to the cell density with OD600 about 0.8, and then placed under different conditions for expression of PEL-SLO. (A) At different temperatures (15 - 37°C), expressions were performed under the induction of 0.6 mM IPTG at 200 rev/min for 36 hours; (B) expressions were induced by different concentrations of IPTG (0.05 - 1.0 mM) at 25°C and 200 rev/min for 36 hours; (C) expressions were performed at 25°C at 200 rev/min under the induction of 0.6 mM IPTG for different lengths of time.
4.2. Purification of Recombinant Protein
Figure 2 demonstrates the result of the purification of the PEL-SLO fusion protein. The predicted molecule weight of the SLO fusion protein was 91.7 Kd. The best purification conditions were the binding buffer (10 mmol/L imidazole, 500 mmol/L NaCl, 20 mmol/L Tris-HCl, pH 7.9), the washing buffer (60 mmol/L imidazole, 500 mmol/L NaCl, 20 mmol/L Tris-HCl, pH 7.9), and 3 elution buffers (100, 200 and 500 mmol/L imidazole, 500 mmol/L NaCl, 20 mmol/L Tris-HCl, pH 7.9). When the concentration of 10 mmol/L imidazole in binding buffer was used, the most effective combination of fusion protein and Ni2+-nitrilotriacetate-agarose column was reached. The most impurities and little goal protein were washed when the 60 mmol/L imidazole concentration in washing buffer was used. Elution was applied by 3 steps with 3 elution buffers (100 mmol/L imidazole, 500 mmol/L NaCl, 20 mmol/L Tris-HCl, pH 7.9; 200 mmol/L imidazole, 500 mmol/L NaCl, 20 mmol/L Tris-HCl, pH 7.9; 500 mmol/L imidazole, 500 mmol/L NaCl, 20 mmol/L Tris-HCl, pH 7.9).
4.3. Hemolysis Characterization of the PEL-SLO Fusion Protein
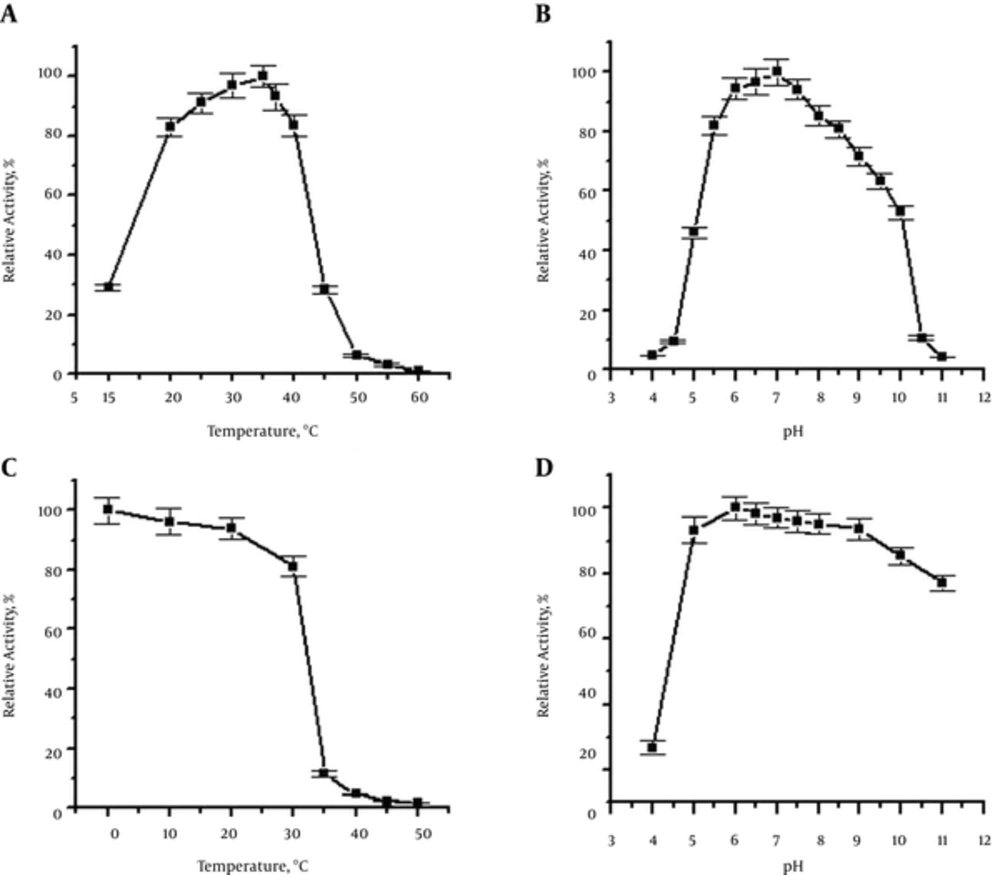
The molecular mass of the recombinant PEL-SLO fusion protein was about 91.7 kDa, determined by SDS-PAGE (Figure 2), which was identical to the prediction with Dnaman soft (http://www.lynnon.com). The calculated pI of the recombinant fusion protein was 5.39. For sheep red cells, the optimum reaction conditions for the recombinant fusion PEL-SLO protein activity were 30 - 40°C (Figure 4A) and pH 6.5 (Figure 4B). Temperature stability test revealed that SLO fusion protein exhibited 80% approximate activity at 0 - 30°C and pH 7.0 for 1 hour (Figure 4C), whereas pH stability test revealed that the fusion protein retained about 80% activity over the range of pH 5.0 - 11 at the most stable temperature for 1 hour (Figure 4D). The hemolysis activity test showed that the specialty of the purified new SLO fusion protein reached 1 × 107 HU/mg.
A, Temperature dependence of the hemolysis activity; B, pH dependence of the hemolysis activity; C, thermo stability of the fusion protein; D, pH stability of the fusion protein. The highest hemolysis activity was designated as 100% in experiments of pH and temperature dependence. The activity of the toxin without pre-incubation was designated as 100% in experiments of temperature and pH stability.
5. Discussion
5.1. The Reason of PEL Promoting the SLO Biosynthesis
The yield of the SLO fusion protein was 600 mg/L culture, and the yield of purified SLO fusion protein was 100 mg/L culture. This yield was about 5 fold of what was previously reported (17, 18). The recombinant mature SLO (containing 32 - 571 amino acids) fused with His-tag was expressed in E. coli, and the yield of the SLO fusion protein was 10 - 30 mg/L culture (16). Velazquez et al. (17) expressed the fusion protein of SLO (78 - 571 acid amino sequence) with GST in E. coli, and its purified SLO yield was 1.5 mg/L. The recombinant mature SLO was expressed as another fusion protein with a thioredoxin and a C-terminal hexahistidine tag, and the yield of SLO fusion protein yield was 124 mg/L medium according to SLO activity concentration (2000 HU/mL cell suspension) and specific activity (16102 U/mg) (15). The reason for PEL promoting the SLO biosynthesis might be reduction in the SLO’s physiological toxicity and the growth inhibition effect on E. coli (data not published).
5.2. The Pectate Lyase Could Strengthen the Hemolysis Activity of SLO
Specific hemolysis activity of purified PEL-SLO was 1 × 107 IU /mg, which was higher than that of native SLO (32 - 571 amino acid sequence) and was about 10 fold of native SLO. Specific hemolysis activity of purified native SLO has been reported as 8 × 105 HU/mg of protein (4). A similar level was also published for recombinant SLO (32 - 571 amino acid sequence) (22). The mature SLO (contain 32 - 571 amino acid) fused with His-tag had a specific activity (748,000 HU/mg protein) and its activity was identical to mature natural or recombinant SLO (16). Our result revealed that PEL fusion tag could strengthen the activity of SLO, and this result was identical to the SLO fusion protein (78 - 571 acid amino sequence) with GST, which had a specific hemolysis activity of 1 ×108 HU/mg (17). Thioredoxin was also used as SLO fusion tag, but the effect of thioredoxin on SLO activity was different from PEL and GST, and its specific activity was only 16102 HU/mg (18).
5.3. Effect of Pectate Lyase on the Antigenicity of Streptolysin O
In the study, recombinant SLO had antigenicity, but recombinant PEL-SLO did not show antigenicity (not published). The antigenicity of proteins was determined by antigenic determinants and the antigenic determinants of the proteins had 2 types of sequential epitope (24, 25) and conformation epitope (26). The research showed that the purified PEL-SLO fusion protein had a specialty of 1 × 107 HU/mg, about 10 fold of natural SLO. Thus, the conformation of PEL-SLO should be right; the effect of pectate lyase on the antigenicity of SLO should belong to the steric effect resulting from pectate lyase although no researches showed SLO had any sequential epitope or conformation epitope. In this study, the pectate lyase was attached to the N-end of SLO fragment, so the antigenic determinant of SLO might be near the N-end of SLO fragment. Conformation epitope might be more easily affected by steric than sequential epitope (27), and the epitope of SLO might be conformation epitope (28-30).
5.4. Conclusion
The yield of the fusion protein of PEL-SLO in E. coli was 600 mg/L culture, and the yield was about 5 fold of the previously reported yield. Specific hemolysis activity of purified PEL-SLO was 1 × 107 IU /mg, which was higher than that of native SLO (32 - 571 amino acid sequence) and was about 10 fold of native SLO. The PEL-SLO fusion protein could meet the increasing need of SLO applications better in animal cell biology and medicine science.