1. Background
The COVID-19 pandemic began with the emergence of the SARS-CoV-2 virus in Wuhan, China, in late 2019 (1). Initially presenting as a pneumonia-like illness, it quickly spread across the globe. Initially referred to as 2019-nCoV, the virus was later renamed SARS-CoV-2 by the International Committee on Taxonomy of Viruses (ICTV), while the disease it caused was designated COVID-19 by the World Health Organization (WHO) (2). The rapid spread of the virus led to its classification as a Public Health Emergency of International Concern (PHEIC) and, by March 2020, a global pandemic. By June 2022, the WHO reported more than 532 million confirmed COVID-19 cases and over 6.3 million deaths worldwide, illustrating the virus’s profound global impact (3).
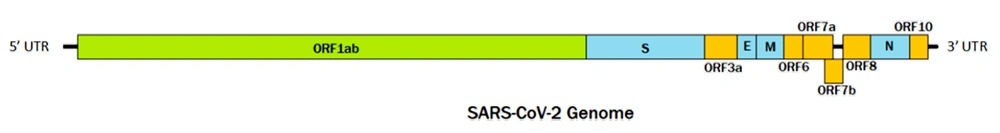
SARS-CoV-2, a novel member of the Betacoronavirus genus and Sarbecovirus subgenus, shares key genetic and structural characteristics with its close relative, SARS-CoV. Both viruses are single-stranded, positive-sense RNA viruses with genomes approximately 29 900 nucleotides in length. They possess similar structural components, including the spike glycoprotein (S), envelope protein (E), membrane glycoprotein (M), and nucleocapsid phosphoprotein (N). Both viruses also encode several accessory proteins, though the number and functions of these proteins may vary between the two. The genomic layout of SARS-CoV-2 is illustrated in Figure 1 (4, 5).
Despite their similarities, SARS-CoV-2 and SARS-CoV exhibit key differences. A notable distinction lies in their origins and natural reservoirs. SARS-CoV is believed to have originated in bats, with transmission to humans occurring via intermediary hosts such as civet cats. While the origins of SARS-CoV-2 remain under investigation, bats are also suspected to be involved, with the possibility of other animal intermediaries. Furthermore, although both viruses can cause severe respiratory illness, SARS-CoV-2 has proven to be more transmissible, resulting in a higher number of global cases. Genomic differences, particularly in the regions encoding the S protein—which is critical for viral entry into host cells—also contribute to the distinct characteristics of these viruses.
Variant of interest (VOI) and variant of concern (VOC) are classifications used by the WHO to categorize specific SARS-CoV-2 mutations or lineages based on their potential public health impact. Variants of Interest are those with genetic changes that may influence virus characteristics such as transmissibility, disease severity, or immune evasion, but there is not yet sufficient evidence of a significant effect on these factors. In contrast, Variants of Concern are those that exhibit more notable changes, including increased transmissibility, more severe disease outcomes, or reduced effectiveness of vaccines and treatments, leading to a higher public health risk.
These classifications are crucial for monitoring and responding to the virus's evolution, as they help prioritize research, surveillance, and public health interventions. Identifying and tracking VOIs and VOCs contributes to understanding how the virus is changing over time and how these mutations may affect the effectiveness of current control measures (6, 7). This study focuses on conducting a phylogenetic analysis and a comprehensive genome mutation analysis of SARS-CoV-2 VOIs and VOCs from different regions. Understanding the virus's genetic variability across countries remains essential in managing the ongoing pandemic (8).
2. Objectives
This study aims to investigate the differences in SARS-CoV-2 Variants of Interest and Concern from various geographical regions through phylogenetic construction and mutational analysis.
3. Methods
3.1. Data collection
A total of 100 sequences per variant were obtained from SARS-CoV-2 VOIs and VOCs, resulting in 700 sequences retrieved from the GISAID database between January and December 2021. These sequences were selected from various geographical regions, including Oceania, Europe, Asia, Africa, South America, and North America, with approximately 14 - 16 sequences from each location. The inclusion criteria for this study included: Samples collected from humans, sequences that were complete (a minimum length of 29 000 base pairs), sequences with less than 5% N-bases, and complete metadata such as the date of collection, specimen source, and patient information (age and sex). These data were used to investigate the link between SARS-CoV-2 genomic variation and known epidemiological information. All sequences examined were organized into a Microsoft Excel spreadsheet (Figure 2).
3.2. Multiple Sequence Alignment
The retrieved genome sequences of VOIs and VOCs were grouped based on region and visualized using Jalview 2.11.2. Multiple sequence alignment was performed for each region using the reference genome from NCBI GenBank (NC_045512) and Clustal Omega v1.2.4 with default settings (9). The consensus sequence generated from each group was compiled and named according to the variant and region for phylogenetic analysis.
3.3. Phylogenetic Analysis
Molecular phylogenetic tree construction began by manually trimming the bases in the 5’-UTR and 3’-UTR. The phylogenetic tree was constructed using the maximum likelihood method with the default settings in the IQ-TREE webserver. Ultra-Fast Bootstrapping (UFBoot) and SH-like approximate likelihood ratio tests (SH-aLRT) were performed with 1,000 iterations (10). The resulting phylogenetic tree was visualized using the Interactive Tree of Life v6.5.7 (11).
3.4. Genome Mutation Analysis
Genome mutation mining was conducted using the GISAID CoVSurver web application. The multiple sequence alignment (MSA) sequences were mapped to the reference sequence 'hCoV-19/Wuhan/WIV04/2019' (Accession number NC_045512.2 in NCBI). This analysis generated lists of amino acid substitutions at specific locations in all structural and non-structural (nsp) proteins. The mutation information was compiled and organized in Microsoft Excel for further analysis.
4. Results
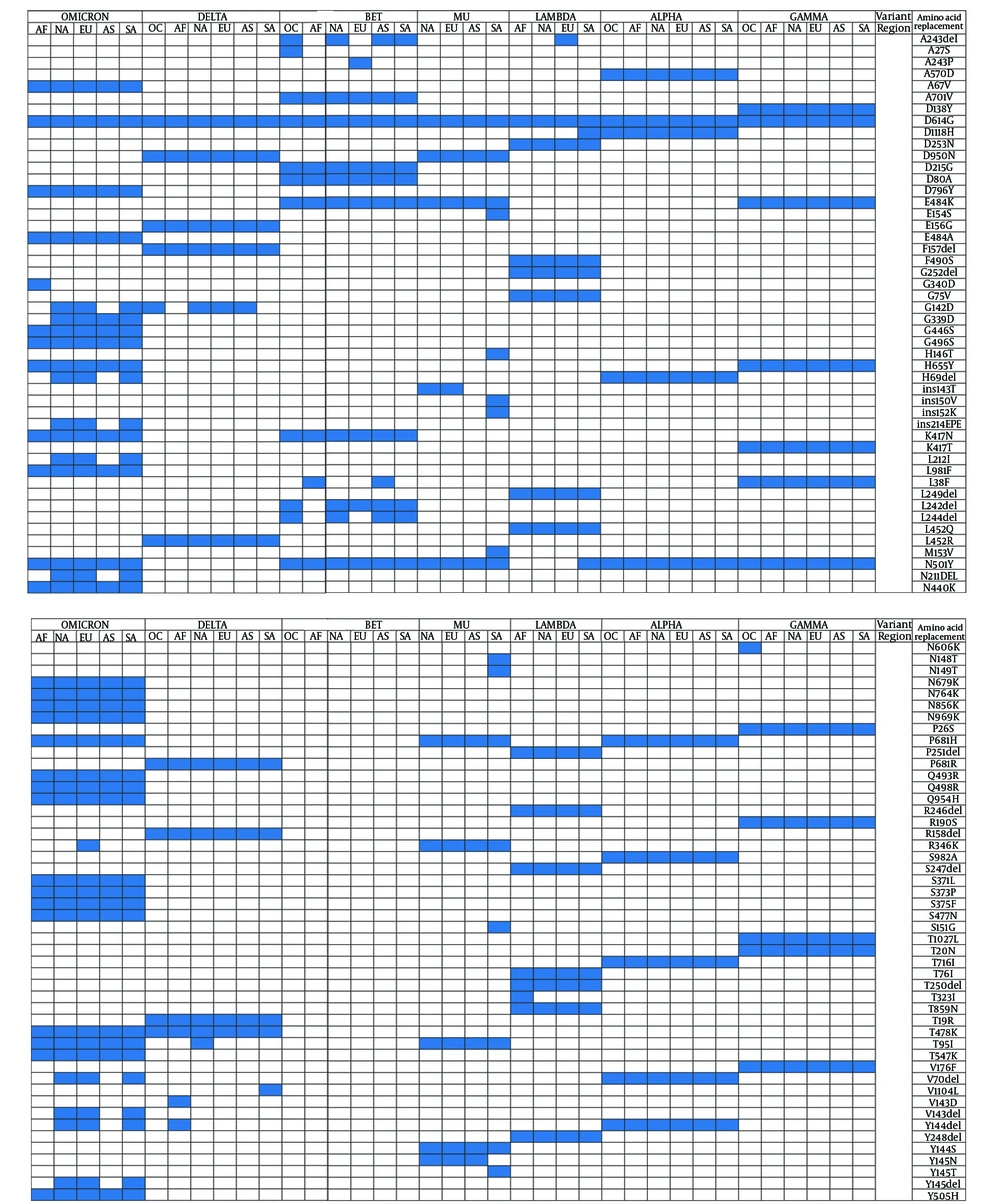
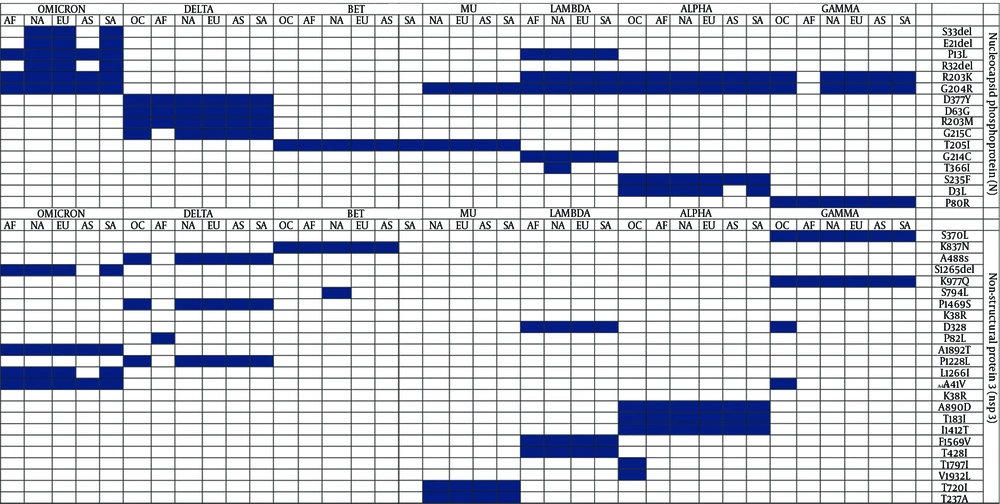
Figures 2 and 3 highlight the protein mutations observed in various SARS-CoV-2 sequences, variants, and regions. Figure 2 identifies 96 mutations, including deletions and insertions, while Figure 3 focuses on 16 mutations in the N protein, non-structural protein 3 (nsp3), and other substitutions. These mutations are shared among VOIs and VOCs, with R203K, G204R, and T205I being common mutations, and T366I specifically found in the VOI Lambda variant from North America. Both Figures 2 and 3 show a combination of novel and shared mutations. Shared mutations suggest genetic changes across multiple variants, indicating common evolutionary pathways or selective pressures.
Notable novel mutations in the S protein include ins143T, ins152K, and ins214EPE, which may impact the structure and function of the S protein, crucial for viral entry into host cells. Alterations in the S protein can affect virus infectivity, transmissibility, and immune recognition (12). Novel mutations in the N protein, such as S33del, E31del, and D3L, can influence viral replication, genome packaging, and the virus’s ability to evade the host immune response (13).
Shared mutations in the S protein, such as A243del and Y144del, indicate common evolutionary pathways among different variants, potentially contributing to increased infectivity and transmission (12). Similarly, shared mutations in the N protein, including R203K, G204R, and T205I, may influence viral replication and packaging (14). Mutations in nsp3, such as A850D, T183I, and I1412T, may affect viral replication and immune evasion, potentially enhancing the virus’s ability to evade host immune defenses and cause disease (15).
In the analysis of S protein mutations, the Omicron variant from South and North America showed the highest mutation count (37/97), followed by Europe (37/97), Asia (28/97), and Africa (26/97). Interestingly, the Alpha variant displayed a similar number of mutation sites across all continents, with 10 mutations each (refer to Figure 2). The highest number of mutations was observed in Omicron sequences from South America and Europe, while the Beta variant had the lowest mutation count, with only one mutation observed across all regions. In non-structural protein 3 (nsp3), Omicron variants exhibited the highest number of mutations, with a total of 18 mutations. North America had the highest number of mutations (19/24), followed by South America (18/24), Europe (18/24), Africa (14/24), Asia (13/24), and Oceania (13/24).
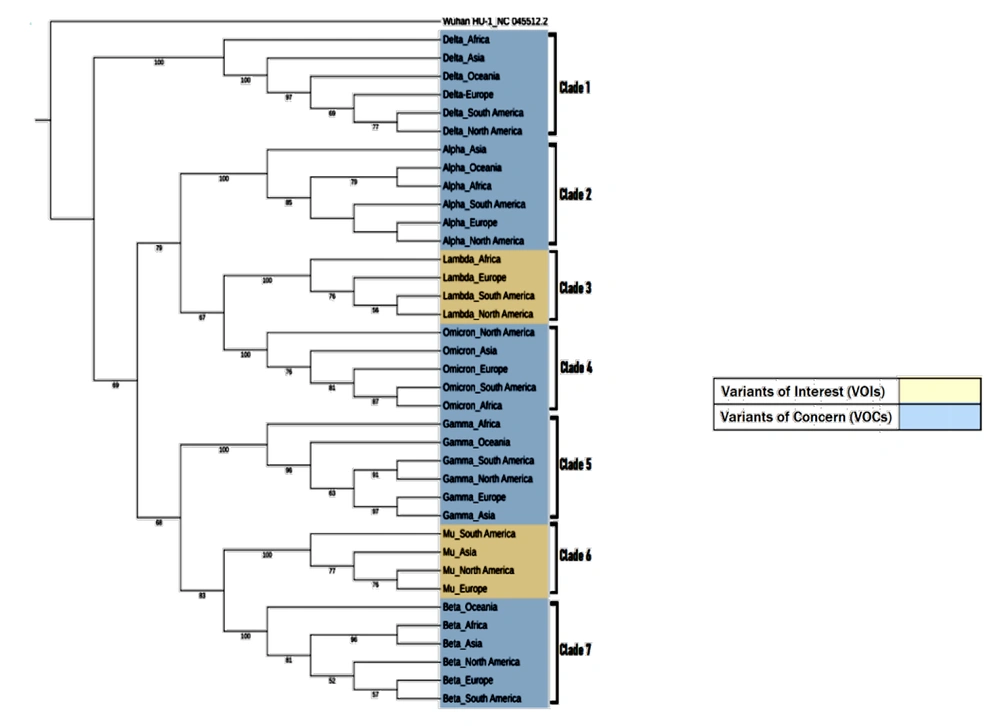
The phylogenetic analysis (Figure 4) of SARS-CoV-2 VOIs and VOCs genomes is based on substitution estimation using the maximum likelihood method, where branching represents evolutionary differences between sequences. A total of 37 sequences were analyzed to represent the full-length genomes of SARS-CoV-2 VOIs and VOCs from various regions. The phylogenetic results showed that sequences of the same variant clustered together as distinct clades, consistent across all VOIs and VOCs. The Delta variant sequences formed a separate cluster (Clade I), clearly distinct from the other VOIs and VOCs.
Maximum-likelihood phylogeny tree based on VOIs and VOCs sequences from different geographical regions: Asia, North America, Europe, Africa, South America, and Oceania. Wuhan/Hu1/2019 (Genbank: NC_045512.2) was used as an outgroup. The peach-colored box highlights VOIs, while the blue box highlights VOCs. Seven clades represent each of the VOIs and VOCs clusters. The red boxes indicate comparisons of sister taxa originating from Africa, while the green boxes show a cluster of closely related sequences from Europe, North America, and South America.
The phylogenetic tree also revealed that the primary root divided into two major groups: Alpha, Lambda, and Omicron (Clades 2, 3, and 4) clustered in one group, while Gamma, Mu, and Beta (Clades 5, 6, and 7) clustered together in another group. A closer analysis of Clade 1's topology showed that the African sequence diverged from those of other regions. This pattern was similarly observed in Clades 3 and 5 (highlighted in the purple boxes). Additionally, comparisons of topologies in Clades 2, 4, 6, and 7 revealed divergence in taxa from Asia, North America, South America, and Oceania, respectively. Furthermore, sister taxa relationships were observed between North America, South America, and Europe in most clades (Clades 1, 2, 3, and 7), as indicated by the red circles.
5. Discussion
This study explores the evolutionary patterns of SARS-CoV-2 variants, emphasizing key mutations and their global distribution in 2021 (16). The phylogenetic analysis reveals that SARS-CoV-2 variants from different regions cluster together based on the mutations they acquired (17). The Delta variant, in particular, branches away from the ancestral virus, hCoV-19/Wuhan/Hu-1/2019 (NC_045512.2), as well as from other VOIs and VOCs, indicating more significant nucleotide differences between them (Refer to Figure 3) (18).
The phylogenetic analysis also revealed close genetic similarities among isolates from Europe, North America, and South America for the Delta, Alpha, Beta, and Lambda variants. The Alpha variant, which originated in the United Kingdom (Europe), was first identified in São Paulo, South America, following an unusual PCR result during routine SARS-CoV-2 diagnosis (18). The genome obtained from the patient formed a cluster strongly supported by 85% bootstrapping of 10 sequences, 60% of which originated in the United Kingdom (Europe). This aligns with the travel history of a close contact of the patient, an asymptomatic family member with SARS-CoV-2, who had traveled from Italy to the United Kingdom and then from London to São Paulo. It is likely that multiple introductions of the Alpha variant occurred in São Paulo, contributing to its prolonged transmission due to the city's significant economic, transportation, and communication networks (19). This supports the conclusion by (20) that travel was a key factor in importing variations and subsequently contributing to infections, illustrating the role of globalization in the spread of the SARS-CoV-2 pandemic. Hence, the Alpha variant from Europe, North America, and South America showed close genetic similarities among the isolates.
Similarly, the South American Lambda variant (sublineage C.37.1) displayed close relationships with samples from North America and Europe, characterized by the presence of spike mutations such as Q675H in North American samples and R21I and T572I in European samples. The Q675H mutation was also found in Lambda variants from South America, while the V826L mutation present in both European and North American Lambda variants further highlighted their close evolutionary relationships (21). Additionally, (22) described genomic analyses during a Beta variant outbreak in Canada (North America), confirming that all B.1.351 genomes were closely related, with fewer than or only two single nucleotide polymorphisms between the sequences.
Following the discovery of the Delta variant in India, it rapidly spread worldwide, with the United Kingdom predominantly associated with the B.1.617.2 lineage. Common mutations found in most B.1.617.2 sequences include T19R, G142D, R158G, L452R, T478K, D614G, P681R, D950N, and deletions at positions 156-157. The B.1.617.2 lineage continues to evolve, with concerning mutations such as K417N emerging in the AY.1/B.1.617.2.1 sub-lineage (23). After the integration of the parent B.1.617.2 lineage, 245 lineages were categorized under AY in the Pango nomenclature system. The AY.44 and AY.103 lineages dominated in California, while AY.20 and AY.26 were prevalent in Mexico. According to (24), the most common Delta-related variants detected in Brazil were AY.99.2, AY.43, AY.101, AY.34.1, AY.43.1, AY.43.2, AY.46.3, AY.100, AY.99.1, and AY.36. Additionally, T19R, T95I, E156G, DEL157/158, L452R, T478K, D614G, Q677H, P681R, D950N, V1104L, and L1265F were frequently reported mutations across more than half of the sequences from these lineages. Thus, the common mutations found in the Delta variant across Europe, North America, and South America suggest close genetic relationships among these genomes.
As mentioned in the results, mutations were observed in nsp, S, E, M, N, and accessory proteins. A study by (25) identified a P314L amino acid mutation caused by the nucleotide change at position 3037 (C3037T), affecting nsp12 and the viral RNA-dependent RNA polymerase. The P314L mutation in nsp12 may enhance viral replication, increasing the virus's transmissibility and infectiousness. Another notable mutation was C14408T, which targets nsp3 (a viral predicted phosphoesterase) at position 14408. According to (26), the most prevalent mutations in nsp3 were P1228L, P1469S, and A488S. In the P1228L mutation, proline (HΦ∼13.5) was replaced by leucine (HΦ∼16.0), leading to reduced stability due to increased hydrophobicity. This mutation, located in the α-helical region of nsp3 within the cytoplasm, may decrease the protein's stability at the P1228L site. In the P1469S mutation, proline (HΦ∼13.5) was replaced by serine (HΦ∼3.0), a hydrophilic amino acid, potentially affecting the protein's stability. A study by (25) suggested that both the P1228L and P1469S mutations in nsp3 may negatively impact protein binding, as these mutations occur near the protein sequence terminal, which is essential for linking nsp3 to nsp4.
Additionally, the A488S mutation replaced alanine (HΦ∼11.0) with serine (HΦ∼3.0), likely increasing the stability of nsp3 in this region. However, this mutation also affected the SUD domain of nsp3, which may influence binding selectivity for G-quadruplexes (G4). This characteristic could play a role in the formation of the replication or transcription viral complex (RTC). Therefore, it is speculated that the A488S mutation may enhance nsp3 stability and strengthen the interaction between the SUD domain and G4s, thereby improving RTC functionality.
The SARS-CoV-2 S proteins have undergone mutations, including changes at glycosylation sites. One of the most prevalent mutations is D614G, which has been shown to significantly enhance viral infectivity (27). The highest density of mutations is located at the S protein's protease cleavage site. These alterations may benefit the virus by allowing it to undergo proteolytic cleavage by various host enzymes, aiding its survival during evolution. Additionally, the spike protein can undergo mutations at multiple sites, with a single site sometimes associated with more than one mutation. To date, the D614G mutation is the only one consistently observed in S proteins across all continents. Mutations near the receptor-binding domain (RBD) that are close to the ACE-2 receptor may impact the shape and charge of the protein near the interaction interface. However, despite these alterations in the S protein's RBD, the virus remains capable of inducing infection (28).
Furthermore, the N501Y mutation, which has been identified in all VOIs and VOCs except Delta, is a S glycoprotein mutation associated with increased viral transmission. This is due to the enhanced binding affinity with the host receptor, ACE2, by slowing the dissociation rate from the receptor (29). Additionally, the K417T mutation in the Gamma variant and the K417N mutation in the Beta and Omicron variants are notable for causing conformational changes in the S protein, contributing to antibody escape (30). Moreover, analysis by (31) indicated that the L452R mutation, found only in the Delta variant's S protein, is associated with increased infectivity and transmission, as it enhances ACE2 binding at the furin cleavage site.
The largest accessory protein in SARS-CoV-2, ORF3a, activates the innate immune receptor NLRP3 inflammasome, triggering host inflammatory responses. This leads to an uncontrolled release of pro-inflammatory cytokines and other mediators, contributing to a cytokine storm—a clinical hallmark of SARS-CoV-2 pathogenesis (32). As projected by (33), mutations in ORF3a may result in the loss of B cell epitopes, thereby affecting ORF3a's antigenicity. Variations in ORF3a could potentially intensify the host immune response, leading to different severities of COVID-19 among individuals, as ORF3a is predicted to interact with host signaling pathways (34). The ORF8 locus in SARS-CoV-2 is highly prone to mutations, including deletions, stop codon changes, and point mutations, with L84S being one of the most frequent. These mutations, particularly deletions, have been associated with milder symptoms and reduced infection severity due to a more effective immune response (35).
Mutations play a crucial role in shaping the clinical outcomes and healthcare response, influencing viral infectivity and transmissibility, as observed with SARS-CoV-2 (36). Key mutations, such as E484Q in the Delta variant and D614G, have been linked to increased infection rates, presenting challenges for public health efforts. These mutations also impact the virus's ability to evade immune responses, potentially affecting the efficacy of monoclonal antibodies and vaccines. Understanding these mutations is essential for developing effective treatments, particularly those targeting virus-host protein interactions (37). Additionally, deletions in regions such as ORF7b have been reported globally, underscoring the need for ongoing research to evaluate their effects on viral fitness and pathogenicity.
5.1. Conclusions
In summary, the ongoing mutation and evolution of SARS-CoV-2 have given rise to various variants with distinct genetic profiles, including mutations in key proteins such as nsp, S, E, M, N, and accessory proteins. These mutations have been linked to changes in infectivity, transmissibility, and potential immune response, all of which influence the clinical progression of COVID-19. Understanding these variants and the specific mutations they carry is critical for developing effective diagnostic, therapeutic, and preventive strategies. Further research, particularly in the field of in silico modeling, is needed to fully grasp the implications of these mutations and their potential impact on public health.