1. Background
Ventilator-associated pneumonia (VAP) is a common nosocomial infection (NI) in critically ill patients. It is associated with a high mortality rate, reaching up to 33% - 50% in high-risk hospital wards, such as intensive care units (ICUs) (1). Acinetobacter baumannii constitutes the ninth most common nosocomial pathogen, reported by the national healthcare safety network (2). It is a bacterium characterized by rapid development of resistance to the majority of antimicrobials and is associated with lower respiratory tract infections in critically ill patients (1, 3-5). This pathogen is responsible for nearly 5% - 10% of VAP cases in ICUs and 61% of VAP-associated mortality in these settings (1, 6). The most common concern among clinicians is the intrinsic resistance of A. baumannii isolates to common antibiotics used for pneumonia treatment (7). Although several outbreaks have been reported in European countries, A. baumannii is known to be endemic to North Africa and Middle East countries including Iran (1, 8). The mechanism of drug resistance acquisition in A. baumannii includes acquisition of mobile genetic elements, such as plasmids, transposons, and integrons from the surroundings (4, 9).
Extended-spectrum beta-lactamase (ESBL)-producing Acinetobacter species are recognized as a major health problem worldwide, considering the limited antibiotic treatment options. ESBL genes are often associated with carbapenem and aminoglycoside resistance genes (10). In addition, integrons may contribute to beta-lactamase overproduction and dissemination (11); also, carrying integrons facilitate the spread of resistance genes among bacteria (12). Epidemiological studies are one of the basic strategies to manage and quickly respond to outbreaks through reducing mortality, changing infection-control settings, and decreasing the incidence of infections and healthcare costs.
There is a significant gap in knowledge about device-associated infections in Iran, especially VAP caused by A. baumannii. Therefore, knowledge about local resistance patterns is essential for exploring feasible alternatives for the management of A. baumannii-associated VAP in developing countries such as Iran with limited resources.
2. Objectives
In the present study, we aimed to assess the presence of ESBL and integron genes in A. baumannii isolates from patients with VAP admitted to the ICUs of 18 hospitals in North of Iran.
3. Methods
3.1. Bacterial Isolation
In this cross sectional study initiated in 2014, nonrepetitive A. baumannii isolates were collected from patients with microbiologically confirmed VAP. This study was approved by the ethics committee of Mazandaran University of Medical Sciences (code, 879; date, July 9, 2014). Ventilator-associated pneumonia was indicated in a mechanically ventilated patient, with chest radiograph showing new or progressive infiltrates, cavitation, consolidation, or pleural effusion at 48 hours after hospitalization.
The inclusion criteria were as follows: 1) onset of purulent sputum or changes in the sputum; and 2) cultured organisms from blood samples or specimens obtained via tracheal aspirate analysis, bronchoalveolar lavage, or biopsy (13). Sampling was carried out over 1 year. All patients with ventilation for at least 48 hours were assessed daily for evidence of ventilator-associated NIs. On the other hand, patients with chronic mechanical ventilation were excluded. The isolates were identified at species level, using standard biochemical tests and microbiological methods (14, 15).
3.2. Antimicrobial Susceptibility Test
Antimicrobial susceptibility testing was performed, using broth microdilution method. The minimum inhibitory concentrations (MICs) were determined according to the standard protocol of the clinical and laboratory standards institute (CLSI). The antimicrobials included amikacin, ciprofloxacin, imipenem, gentamicin, ceftazidime, tobramycin, piperacillin/tazobactam, cefepime, colistin, and cotrimoxazole. The antibiotics were purchased from Sigma Co. (Germany) (16). The XDR Acinetobacter species were defined as isolates resistant to 3 classes of antimicrobials and insensitive to carbapenem (17).
3.3. Phenotypic ESBL Detection
ESBL-producing A. baumannii was detected, using the double-disk synergy test. The presence of ESBL was assayed, using the following antibiotic disks: cefotaxime (30 μg), cefotaxime/clavulanic acid (30/10 μg), ceftazidime (30 μg), and ceftazidime/clavulanic acid (30/10 μg) (MAST, UK). Escherichia coli ATCC 25922 strains served as the positive controls (18).
3.4. DNA Extraction and Polymerase Chain Reaction (PCR) Assay
DNA extraction was carried out, using a commercial standard kit (DNA Zist, Iran). ESBL-positive A. baumannii isolates were screened for CTX, VEB, GES, SHV, int1, and int2 genes via PCR amplification. The PCR primers and annealing temperature are presented in Table 1. After PCR reactions, the PCR products were subjected to 2% gel electrophoresis for 50 minutes (voltage, 70). The results were evaluated under UV light on a UV transilluminator. Also, Klebsiella pneumoniae 7881 (CTXM), K. pneumoniae 7881 (containing SHV), Pseudomonas aeruginosa ATCC 27853 (VEB-1), and K. pneumoniae (GES) were used as the positive controls for ESBL-producing genes. In addition, E. coli 96K062 was used as the positive control for class 1 and 2 integrons (int1 and int2). A non-ESBL-producing strain (E. coli ATCC 25922) was used as the negative control.
| Target Gene | Primer Sequences (5’ - 3’) | Annealing Temperature, °C | Amplicon Size, bp | Reference |
|---|---|---|---|---|
| CTX | TTTGCGATGTGCAGTACCAGTAA | 53.1 | 593 | (19) |
| CGATATCGTTGGTGGCATA | ||||
| VEB | CGACTTCCATTTCCCGATGC | 54.9 | 585 | (20) |
| GGACTCTGCAACAAATACGC | ||||
| GES | ATGCGCTTCATTCACGCAC | 55 | 846 | (21) |
| CTATTTGTCCGTGCTCAGG | ||||
| SHV | AAGATCCACTATCGCCAGCAG | 60 | 231 | (22) |
| ATTCAGTTCCGTTTCCCAGCGG | ||||
| INT1 | CAGTGGACATAAGCCTGTTC | 55 | 160 | (23) |
| CCCGAGGCATAGACTGTA | ||||
| INT2 | TTGCGAGTATCCATAACCTG | 58 | 288 | (24) |
| TTACCTGCACTGGATTAAGC |
3.5. Statistical Analysis
For statistical analysis, descriptive tests were calculated using SPSS version 16.
4. Results
Out of 205 patients with NI admitted to the ICUs of 18 hospitals, 29 had VAP caused by ESBL-producing A. baumannii. In total, 19 (65.5%) patients were male and 10 (34.5%) were female. The average age of the participants was 62.16 ± 17.76 years. The susceptibility patterns of ESBL-producing A. baumannii to different categories of antibiotics are presented in Table 2.
| Antibiotics | Resistant | Intermediate | Sensitive |
|---|---|---|---|
| Aminoglycosides | |||
| Amikacin | 23 (79) | 2 (6.9) | 4 (13.8) |
| Gentamicin | 27 (93.1) | 2 (6.9) | 0 |
| Tobramycin | 18 (62.1) | 7 (24.1) | 4 (13.8) |
| Fluoroquinolones | |||
| Ciprofloxacin | 24 (82.8) | 3 (10.3) | 2 (6.9) |
| Carbapenems | |||
| Imipenem | 16 (55.2) | 7 (24.1) | 6 (20.7) |
| Penicillin + β-lactamase inhibitors | |||
| Piperacillin/tazobactam | 28 (96.6) | 1 (3.4) | 0 |
| Cephalosporins | |||
| Ceftazidime | 26 (89.7) | 2 (6.9) | 1 (3.4) |
| Cefepime | 26 (89.7) | 2 (6.9) | 1 (3.4) |
| Polypeptides | |||
| Colistin | 10 (34.5) | 13 (44.8) | 6 (20.7) |
| Sulfonamide + trimethoprim | |||
| Cotrimoxazole | 26 (89.7) | 1 (3.4) | 2 (6.9) |
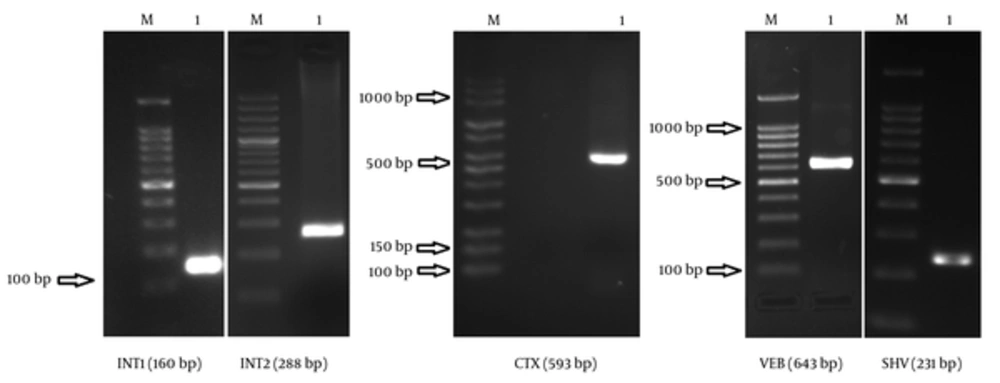
Based on the findings, 19 (65.51%) strains were susceptible to only 1 or 2 antibiotic categories. The prevalence of CTX, VEB, and SHV genes in ESBL-producing A. baumannii was 34.5%, 17.2%, and 96.6%, respectively; however, none of the isolates contained GES genes. PCR detection of integrase genes showed that 79.3% of the isolates had class 1 integrons, while 10.3% had class 2 integrons. Figure 1 illustrates Agarose gel electrophoresis of strains containing VEB, SHV, CTX, int1, and int2 genes.
The antibiotic susceptibility pattern of A. baumannii, containing ESBL-producing genes and integrons, is shown in Table 3. The coincidence of isolates with different ESBL gene types and class 1 integrons is presented in Table 4. One strain contained 3 ESBL genes (VEB, CTX, and SHV), while 14 strains had 2 ESBL genes (10 strains, CTX and SHV; 4 strains, VEB and SHV). Based on the findings, 14 strains had only 1 ESBL gene (SHV). The coincidence of isolates containing different types of ESBL genes and class 1 integrons was approximately 72% - 100%, which is statistically significant (P > 0.05).
| Antibiotics | SHV | VEB | CTX | INT1 | INT2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| NEG (N = 1) | POS (N = 28) | NEG (N = 24) | POS (N = 5) | NEG (N = 19) | POS (N = 10) | NEG (N = 6) | POS (N = 23) | NEG (N = 26) | POS (N = 3) | |
| Amikacin | ||||||||||
| R | 1 (100) | 22 (78.57) | 19 (79.16) | 4 (80) | 14 (73.68) | 9 (90) | 6 (100) | 17 (73.91) | 20 (76.92) | 3 (100) |
| I | 0 | 2 (7.14) | 1 (4.16) | 1 (20) | 2 (10.52) | 0 | 0 | 2 (8.69) | 2 (7.69) | 0 |
| S | 0 | 4 (14.28) | 4 (16.66) | 0 | 3 (15.78) | 1 (10) | 0 | 4 (17.39) | 4 (15.38) | 0 |
| Ciprofloxacin | ||||||||||
| R | 1 (100) | 23 (82.14) | 19 (79.16) | 5 (100) | 15 (78.94) | 9 (90) | 5 (83.3) | 19 | 22 (84.61) | 2 (66.6) |
| I | 0 | 3 (10.71) | 3 (12.5) | 0 | 3 (15.78) | 0 | 1 (16.6) | 2 (8.69) | 2 (7.69) | 1 (33.3) |
| S | 0 | 2 (7.14) | 2 (8.33) | 0 | 1 (5.26) | 1 (10) | 0 | 2 (8.69) | 2 (7..69) | 0 |
| Imipenem | ||||||||||
| R | 1 (100) | 15 (53.57) | 14 (58.33) | 2 (40) | 13 (68.42) | 3 (30) | 5 (83.3) | 11 (47.82) | 14 (53.84) | 2 (66.6) |
| I | 0 | 7 (25) | 6 (25) | 1 (20) | 3 (15.78) | 4 (40) | 0 | 7 (30.43) | 6 (23.07) | 1 (33.3) |
| S | 0 | 6 (21.42) | 4 (16.66) | 2 (40) | 3 (15.78) | 3 (30) | 1 (16.6) | 5 (21.73) | 6 (23.07) | 0 |
| Gentamicin | ||||||||||
| R | 1 (100) | 26 (92.85) | 23 (95.83) | 4 (80) | 17 (89.47) | 10 (100) | 6 (100) | 21 (911.3) | 24 (92.30) | 3 (100) |
| I | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 (7.69) | 0 |
| S | 0 | 2 (7.14) | 1 (4.16) | 1 (20) | 2 (10.52) | 0 | 0 | 2 (8.69) | 0 | |
| Ceftazidime | ||||||||||
| R | 1 (100) | 25 (89.28) | 23 (95.83) | 3 (60) | 16 (84.21) | 10 (100) | 6 (100) | 20 (86.95) | 23 (88.46) | 3 (100) |
| I | 0 | 2 (7.14) | 1 (4.16) | 1 (20) | 2 (10.52) | 0 | 0 | 2 (8.69) | 2 (7.69) | 0 |
| S | 0 | 1 (3.57) | 0 | 1 (20) | 1 (5.26) | 0 | 0 | 1 (4.34) | 1 (3.84) | 0 |
| Tobramycin | ||||||||||
| R | 1 (100) | 17 (60.71) | 17 (70.83) | 1 (20) | 13 (68.42) | 5 (50) | 5 (83.3) | 13 (56.52) | 17 (65.38) | 1 (33.3) |
| I | 0 | 7 (25) | 3 (12.5) | 4 (80) | 5 (26.31) | 2 (20) | 1 (16.6) | 6 (26.08) | 5 (19.23) | 2 (66.6) |
| S | 0 | 4 (14.28) | 4 (16.66) | 0 | 1 (5.26) | 3 (30) | 0 | 9 (39.13) | 4 (15.38) | 0 |
| Piperacillin/tazobactam | ||||||||||
| R | 1 (100) | 27 (96.42) | 23 (95.83) | 5 (100) | 18 (94.73) | 10 (100) | 6 (100) | 22 (95.65) | 25 (96.15) | 3 (100) |
| I | 0 | 1 (3.57) | 1 (4.16) | 0 | 1 (5.26) | 0 | 0 | 1 (4.34) | 1 (3.84) | 0 |
| S | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cefepime | ||||||||||
| R | 1 (100) | 25 (89.28) | 21 (87.5) | 5 (100) | 16 (84.21) | 10 (100) | 6 (100) | 20 (86.95) | 24 (92.3) | 2 (66.6) |
| I | 0 | 2 (7.14) | 2 (8.33) | 0 | 2 (10.52) | 0 | 0 | 2 (8.69) | 1 (3.84) | 1 (33.3) |
| S | 0 | 1 (3.57) | 1 (4.16) | 0 | 1 (5.26) | 0 | 0 | 1 (4.34) | 1 (3.84) | 0 |
| Colistin | ||||||||||
| R | 1 (100) | 9 (32.14) | 9 (37.5) | 1 (20) | 6 (31.57) | 4 (40) | 2 (33.3) | 8 (34.78) | 9 (34.6) | 1 (33.3) |
| I | 0 | 13 (46.42) | 11 (45.83) | 2 (40) | 9 (47.36) | 4 (40) | 4 (66.6) | 9 (39.13) | 11 (42.3) | 2 (66.6) |
| S | 0 | 6 (21.42) | 4 (16.66) | 2 (40) | 4 (21.05) | 2 (20) | 0 | 6 (26.08) | 6 (23.07) | 0 |
| Cotrimoxazole | ||||||||||
| R | 1 (100) | 25 (89.28) | 22 (91.66) | 4 (80) | 17 (89.47) | 9 (90) | 6 (100) | 20 (86.95) | 23 (88.46) | 3 (100) |
| I | 0 | 1 (3.57) | 0 | 1 (20) | 1 (5.26) | 0 | 0 | 1 (4.34) | 1 (3.84) | 0 |
| S | 0 | 2 (7.14) | 2 (8.33) | 0 | 1 (5.26) | 1 (10) | 0 | 2 (8.69) | 2 (7.69) | 0 |
Abbreviations: NEG, Negative; POS, Positive.
| ESBL Type | Number (N = 29) | Coincidence with Class 1 Integrons (N = 23) | P Value | Number (N = 3) | Coincidence with Class 2 Integrons (N = 2) | P Value |
|---|---|---|---|---|---|---|
| VEB, CTX, and SHV | 1 (3.4) | 1 (100) | ≤ 0.0001 | 0 | 0 | - |
| CTX and SHV | 10 (34.48) | 8 (80) | 0.04 | 1 (33) | 1 (100) | ≤ 0.0001 |
| VEB and SHV | 4 (13.79) | 4 (100) | 0.002 | 0 | 0 | - |
| SHV | 14 (48.27) | 10 (71.42) | 0.1 | 2 (66) | 1 (50) | 1 |
5. Discussion
Based on the findings, ESBL-producing A. baumannii was responsible for 14.15% of VAP cases in our region. Also, XDR ESBL-producing A. baumannii was a major cause of VAP with an incidence of 65.51%. During the study, the outbreak of multidrug-resistant (MDR) A. baumannii was reported in the ICU of Imam Sajjad hospital, Ramsar, Iran and was reserved for patients requiring intensive care (e.g., mechanical ventilation). A total of 13 nonduplicate A. baumannii isolates were collected from the clinical specimens in less than 6 months. All the samples were isolated from VAP patients at the hospital. In total, 84.6% of A. baumannii isolates from the outbreak contained class 1 integrons. The occurrence of 2 this outbreak in Mazandaran Province is consistent with several reports on the extensive spread of MDR A. baumannii, causing multicenter outbreaks in European countries and USA (25-27).
The most remarkable finding of the present study was the high rate of class 1 integrons (79.3%) among ESBL-producing A. baumannii isolates from VAP patients. In this study, class 1 integrons were associated with various ESBL-related genes and played an important role in the development of antibiotic resistance. In this regard, in a study by Rahimzadeh et al., the prevalence of class 1 integrons in ESBL-producing A. baumannii was 73%, which is close to the present finding (28). Moreover, Farajnia et al. reported the prevalence of integrons to be 74% among ESBL-producing A. baumannii isolates (12). The rate of integron positive strains in these studies was close to the present findings. However, it should be noted that in the present study, all ESBL-producing A. baumannii isolates were collected from VAP patients, whereas in studies by Rahimizadeh et al. and Farajnia et al., only 37% and 54% of A. baumannii isolates were from VAP patients, respectively.
Recently, colistin resistance has emerged worldwide due to colistin therapy and nosocomial transmission of resistant strains. Nevertheless, colistin is rarely prescribed or used in our region. The possible reasons for the presence of colistin-resistant pathogens in humans without prior exposure to colistin include cross-resistance between colistin and human cationic antimicrobials (e.g., LL-37 and lysozyme) and acquisition of colistin-resistant bacteria from animal food (this antibiotic is heavily used in veterinary medicine).
Another possibility is the involvement of plasmid-mediated colistin resistance, reported in E. coli and K. pneumoniae. The mcr-1 gene, encoded for phosphoethanolamine transferase, was borne on a mobile plasmid in E. coli and K. pneumoniae and could transfer to Enterobacteriaceae and P. aeruginosa (29-31). Therefore, timely detection and isolation of patients harboring colistin resistance and prevention of treatment failure depend on colistin resistance screening in patients, even those without a history of colistin use is necessary.
Today, imipenem therapy is the gold standard for pneumonia due to A. baumannii. Polymyxins remain an exception with regard to the emergence of resistance to the available antibiotics (32). Colistin and imipenem were found to be more effective antibiotics for the treatment of infections caused by resistant isolates. ESBL-producing A. baumannii showed coresistance to different categories of antibiotics, such as quinolones and aminoglycosides. About 70% of A. baumannii isolates in the present study were resistant to aminoglycosides (33, 34). Although CTX gene is more common in some countries, the most prevalent ESBL gene detected in our study was blaSHV (96.6%). The high rate of SHV genes in the present study could be explained by high resistance to aminoglycosides. Based on the findings, resistance to aminoglycosides among A. baumannii strains was 63% - 93%. The association of blaSHV with aminoglycoside resistance genes, such as aacC1, aphA6, aadA1, and aadB, has been established in the literature.
The incidence of aminoglycoside resistance among SHV-encoding A. baumannii isolates was 60% - 92%. In this regard, Safari et al. and Sharif et al. reported prevalence rates of 58% and 63% for SHV gene, respectively, whereas Alyamani et al. could not detect any SHV genes. Moreover, Huang et al. reported a prevalence of 30%, and Hussain et al. showed that in Pakistan, 40% of A. baumannii isolates carried beta-lactamase-resistant SHV genes (35-38); blaSHV genes could be carried by both chromosomes and plasmids. Consequently, clonal dissemination and horizontal gene transfer by integrons (96% in strains with class 1 integrons vs. 100% in strains with class 2 integrons in the present study) both contribute to the overwhelming prevalence of blaSHV genes in A. baumannii isolates.
CTX β-lactamase, produced by A. baumannii strains, is plasmid-mediated, considering the long duration of survival in hospital settings. The prevalence of blaCTX gene in the present study was 34.5%; also, 34.7% of class 1 integron-positive strains contained blaCTX genes. Also, in this study, resistance among strains with blaCTX genes ranged from 30% to 100%. In this regard, Hakemi et al. reported the prevalence of CTX genes to be 10.7% among A. baumannii isolates from wound samples. On the other hand, Alyamani et al. showed that 81% of A. baumannii isolates from the clinical samples of ICU patients contained CTX genes (36, 39). In consistence with the present findings, Ahanjan et al. reported a prevalence of 31.5% in A. baumannii isolates from patients with wound infections (40); this consistency might be explained by the similarities in study settings.
Moreover, we found that 17.2% of the strains contained VEB genes. According to the literature, A. baumannii, harboring integron-borne VEB-1 (an ESBL), had caused outbreaks in French and Belgian hospitals (20, 41). The prevalence of this gene was reported to be 10% in a study by Farajnia et al., 39.5% in a study by Fallah et al., 26.6% in a study by Fazeli et al. in Iran, and 47.61% in the Unites States (12, 42, 43). The coincidence of different ESBL types, containing VEB genes and class 1 integrons, was significant (P < 0.05). In the present study, the high rate of CTX, SHV, and VEB genes in A. baumannii might have been influenced by mobile genetic elements such as integrons (P < 0.05). According to our researches, in recent years the incidence of antibiotic resistance tragically has exponentially increased in north of Iran (44-52)
5.1. Conclusion
Presence of class 1 and class 2 integrons in A. baumannii has been considerably associated with ESBL strains. The high rate of SHV genes and class 1 integrons highlights the necessity of avoiding aminoglycosides for empirical therapy. Although colistin was the most sensitive antibiotic in XDR strains in our region, due to the presence of patients with A. baumannii in ICUs, colistin resistance screening should be performed before empirical therapy for VAP, even for those without prior exposure to this antibiotic.