1. Background
Staphylococcus aureus causes a wide range of diseases, from benign skin infections to life-threatening systemic infections such as pneumonia, endocarditis, and toxic shock syndrome. Methicillin resistant Staphylococcus aureus (MRSA) are multidrug resistant bacteria that often cause serious problems via nosocomial infections in hospitals. The ability of these bacteria to cause such severe infections is related to numerous virulence factors that allow these bacteria to adhere to surfaces/tissues, avoid or invade the immune system, and cause harmful toxic effects in the host. The most important factors are secreted hemolysins, toxins, extracellular enzymes, anti-inflammatory peptides, adhesions, and immune evasion mechanisms (1, 2).
The agr system plays a central role in the regulation of virulence factors during growth. Agr is a quorum sensing system in S. aureus, which is activated during increase in cell density at transition from exponential phase to the stationary phase growth; there is an up-regulated expression of secreted exo-proteins, such as α-hemolysin and down-regulated expression of cell surface associated virulence factors, such as protein A (3, 4). When S. aureus infects a host, the expression pattern of virulence factors in the host seems to be somewhat different from that in culture media (5).
2. Objectives
This study investigated expression of virulence factors in brain heart infusion (BHI) and BHI broth supplemented with sheep blood (5%) to mimic the in-vivo environment from exponential and stationary phase. Selected virulence genes, including hla (encoding alpha-toxin), are generally activated by agr and spa (encoding protein A), and generally repressed by agr and agrA, as representative of agr system and mecA (encoding resistance of methicillin). This study also investigated the expression of one regulatory gene, RNAIII, the effecter molecule of the accessory gene regulator (agr) system, which is reported to up-regulate toxin genes and down-regulate adhesion genes.
3. Methods
3.1. Bacterial Strains
This study was carried out on 5 S. aureus isolates including 2 MRSA and two methicillin sensitive Staphylococcus aureus (MSSA) isolates, which comprised of one isolate from a healthy carrier patient and one from a clinical sample, in each group, and a COL standard isolate. The isolates were recovered and pure culture was prepared from them. The isolates were confirmed with conventional laboratory techniques, including Gram staining, catalase test, hemolysis on blood agar, and coagulase test.
3.2. Plotting of Growth Curve
For this purpose, dilution of 103 CFU/mL for each of the isolated bacteria in BHI broth and BHI broth containing 5% sheep blood (BHIB) was used and this concentration was considered as 0-time for inoculation. These cultures were incubated at 37°C in 24 test tubes, which were sampled at hourly intervals and were inoculated on Muller Hinton agar medium (Merck, Germany). The number of colonies was counted after a 24-hour incubation at 37°C, and the growth curve was plotted. Also absorbance values at 578 nm (OD 578 nm) were recorded for each suspension at hourly intervals (6, 7). Also amounts of the samples were stored for RNA extraction and Real time polymerase chain reaction (PCR) immediately frozen at -70°C until examination.
3.3. Extraction and Purification of RNA
The RNA was extracted at 3 and 7 hours as the exponential phase and 11 and 15 as the stationary phase in Brain Heart Infusion (BHI) broth and BHI containing 5% sheep blood (BHIB). Total RNA of samples were extracted using RNeasy Mini Kit guidelines (Qiagen, Germany), according to the manufacturer’s instructions. This kit contains guanidine thiocyanate buffer. For very sensitive applications, such as RT-PCR, the RNA product should be treated with DNase to remove small amounts of residual DNA. For this purpose, the RNase-Free DNase set (Qiagene, Germany), according to the manufacturer’s directions, was used. Absence of DNA was confirmed by Polymerase Chain Reaction (PCR) and in order to determine the concentration of RNA extracted from Eppendorf Biophotometer, a wavelength of 260 nm (A260) was used, and the purity of the extracted RNA was identified from about 260 nm to 280 nm (A260 / A280). Integrity and quality of RNA were tested by agarose gel electrophoresis and ethidium bromide staining tests so that two bands of 1.5 (16S) and 2.9 (23S) kb, were observed.
3.4. Real Time Polymerase Chain Reaction and Analysis of Gene Expression
To determine the amount of each gene in extracted RNA, Real-Time PCR was performed using specific primers for each of the 5 genes, and expression levels were normalized to the expression of gyrB as an internal control. Real-Time PCR was carried out using SYBR Green through the One-Step RT-PCR method and with QuantiFast SYBR Green RT-PCR kit (Qiagen, Germany), according to the manufacturer’s instructions. For this purpose an ABI 7300 thermocycler was used (Applied Biosystems). Primers used in this study are presented in Table 1; these were confirmed with the NCBI website using Blast pairing place and length of PCR product.
| Name | Sequence (5 to 3) | Product Size, bp | Reference |
|---|---|---|---|
| agrA (F) | TGA TAA TCC TTA TGA GGT GCT T | 163 | (8) |
| agrA (R) | CAC TGT GAC TCG TAA CGA AAA | ||
| RNAIII (F) | CGA TGT TGT TTA CGA TAG CTT | 146 | (5) |
| RNAIII (R) | CCA TCC CAA CTT AAT AAC CA | ||
| hla (F) | GGG GAC CAT ATG ATA GAG ATT | 154 | (5) |
| hla (R) | TGT AGC GAA GTC TGG TGA AA | ||
| spa (F) | GAT GGT AAC GGA GTA CAT GTC GTT | 163 | (9) |
| spa (R) | TTG CTG GTT GCT TCT TAT CAA CA | ||
| mecA (F) | ACT GCT ATC CAC CCT CAA AC | 163 | (10) |
| mecA (R) | CTG GTG AAG TTG TAA TCT GG | ||
| gyrB (F) | CGC AGG CGA TTT TAC CAT TA | 141 | (11) |
Relative target expression was calculated according to the∆ (∆CT) method (12, 13), in which the fold-change in expression is equal to 2 -∆∆CT. Data are presented as fold-change in transcript levels in BHIB relative to BHI broth and the stationary phase relative to the exponential phase.
(1) ∆ CT = CT,target - CT,reference
(2) ∆∆ Ct = ∆Ct(sample) - ∆Ct(calibrator)
3.5. Statistical Analysis
Each experiment was replicated 3 times and the results were analyzed by the SPSS software through analysis of variance (ANOVA) and Duncan test at a significance level of 0.05.
4. Results
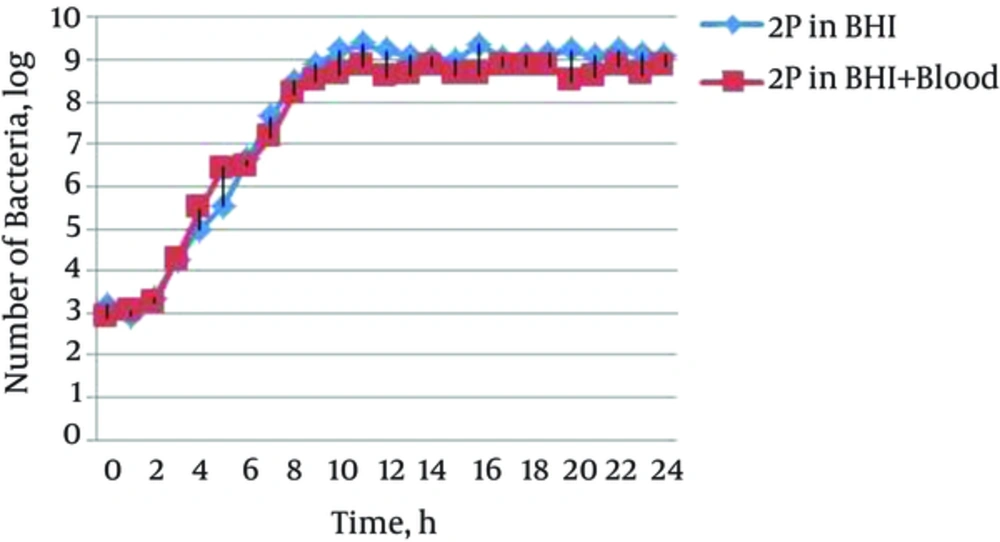
The growth curves of 5 S. aureus isolates in 2 different growth media; including BHI and BHIB were plotted during time of inoculation (0-time) through a 24-hour incubation period. Figure 1 shows the growth curve of a clinical isolate of methicillin-resistant S. aureus in two growth media; BHI and BHIB at 24 hours (Figure 1). The growth curves of other isolates were similar to this isolate, in which the corresponding data are not shown.
The results showed that the beginning of the exponential phase, mid-exponential phase, stationary phase, and mid-stationary phase in isolates and different cultures were at hours 3, 7, 11, and 15, respectively, and there was no significant difference between cultures of BHI and BHI containing blood (BHIB). For this reason, the gene’s expression was evaluated at the mentioned times, and mean CT at 3 and 7 hours was considered as the exponential phase and mean CT at 11 and 15 hours, as the stationary phase.
4.1. Profile of Gene Expression at Different Growth Phases
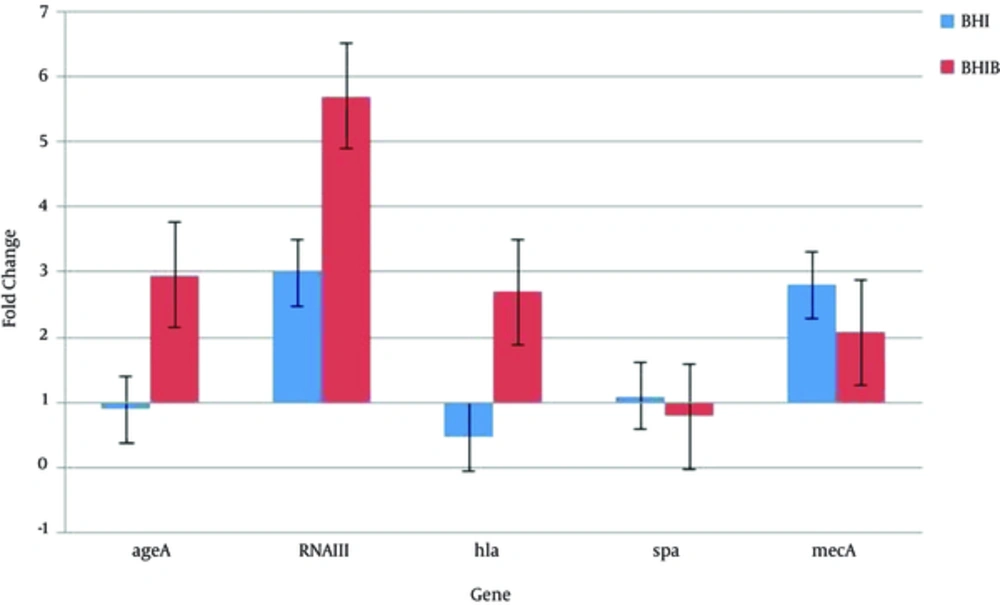
As shown in Figure 2, when considering the mean of 5 isolates consequently, the mean agrA gene expression in BHI broth containing 5% sheep blood (BHIB) showed a 2.95-fold increase in stationary growth phase relative to exponential phase, while its expression in BHI broth decreased during the stationary phase. The RNAIII acts as an effector molecule in the agr system. In both culture conditions, mean expression of this gene in stationary phase was higher than the exponential phase. A 5.7-fold increased expression was found in the BHI broth containing 5% sheep blood (BHIB) and a 2.99-fold increase in BHI. The expression of the hla gene is responsible for synthesis of alpha hemolysin and is one of the most important virulent factors of S. aureus; in medium containing blood (BHIB) a 2.7-fold increase was detected while mean expression of this gene in standard laboratory medium (BHI broth) in stationary growth phase relative to exponential phase was reduced. The spa gene encodes for protein A that is one of the most important bacterial defense proteins against non-specific immune defense. Expression of this gene decreased in medium containing blood (BHIB) and finally, mean expression of gene encoding resistance to methicillin (mecA) in 5 isolates, increased in both environmental conditions (Figure 2).
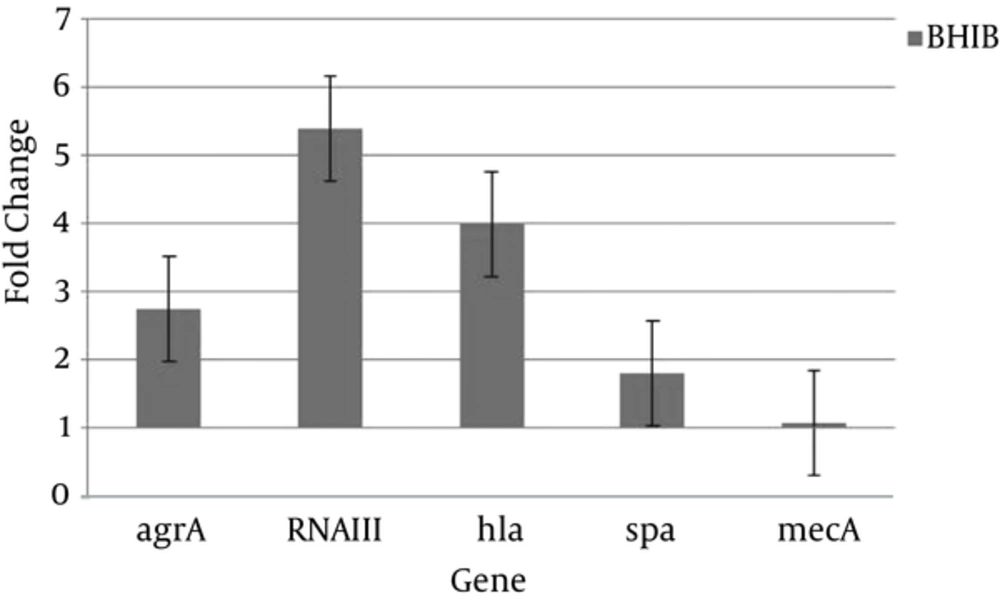
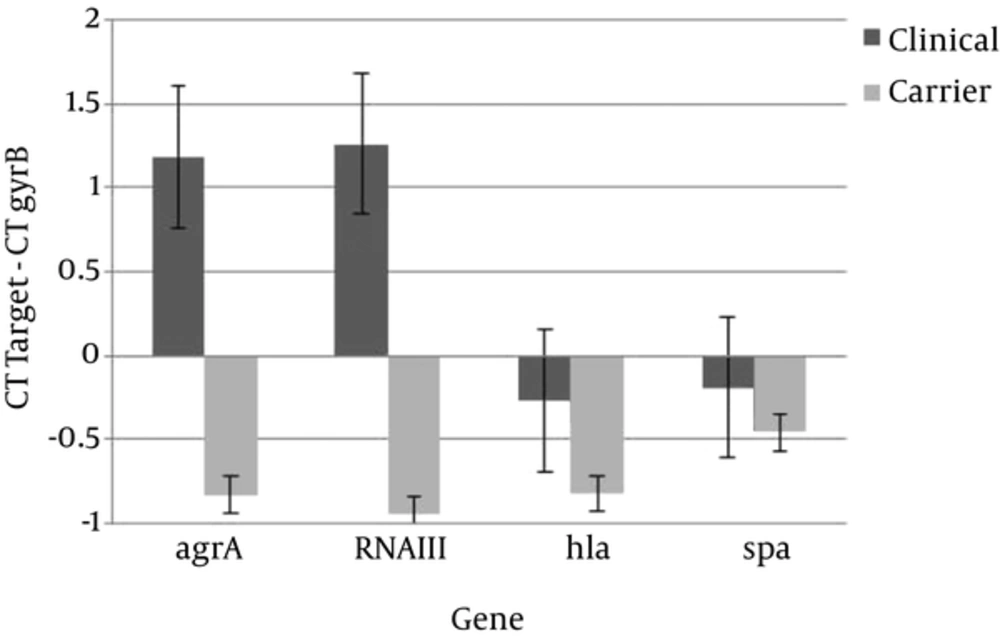
Also Figure 3 shows that regardless of the growth phase, the expression of all studied genes increased in BHI broth containing 5% sheep blood (BHIB) compared to BHI broth. This change was more than other genes for RNAIII gene expression (Figure 3). The other aim of this study was to compare expression levels of virulence and regulatory of genes in colonizing and invasive isolates. Transcription of all of genes was up-regulated in colonizing isolates compared to invasive isolates (Figure 4).
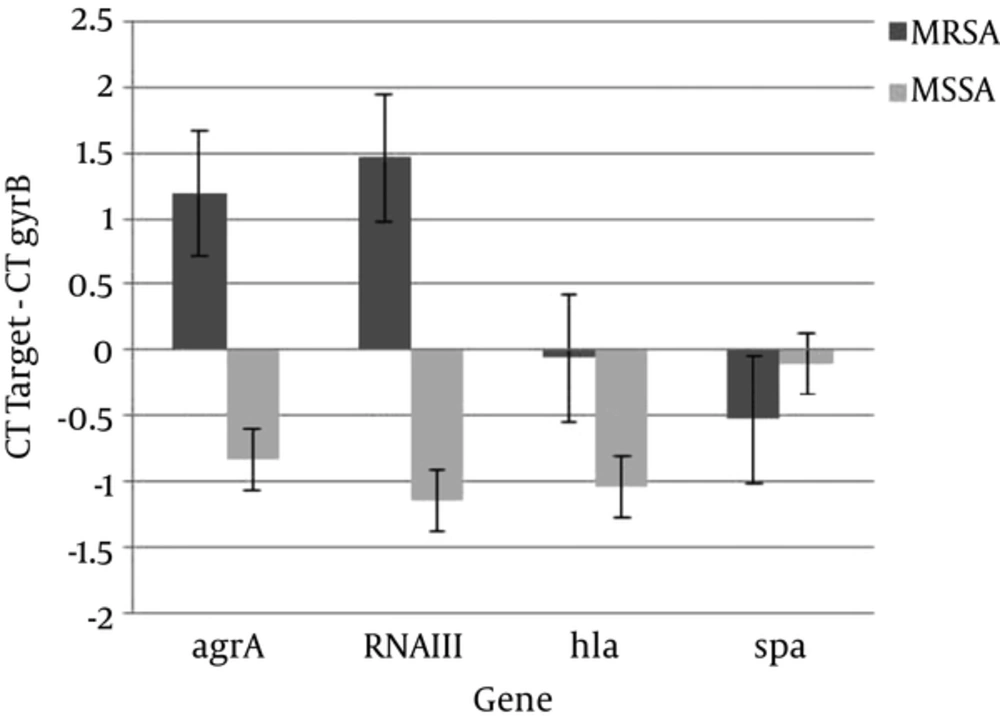
The other aim of this study was to compare expression levels of virulence and regulatory genes in MRSA and MSSA isolates. Transcription of all of the genes except spa, was up-regulated in MSSA isolates compared to MRSA isolates (Figure 5).
5. Discussion
Several studies have implied that BHI broth is a highly nutritious culture and ideal for bacterial growth and does not need adding of blood, as it is not nutritious. Additives such as sheep blood and calf serum cannot increase the growth rate (5, 14, 15). The results showed that the addition of 5% sheep blood in BHI broth could not reduce the generation time of S. aureus or increase growth rate of this bacteria, yet this enrichment compound could enhance expression of virulence genes of S. aureus.
The Agr is a quorum sensing system in S. aureus, which is activated during increase in cell density at transition from exponential phase to stationary phase growth; there is an up-regulated expression of secreted exoproteins, such as α-hemolysin, and down-regulated expression of cell surface-associated virulence factors, such as Protein A (16-18). In the current study, the agr system in the BHI broth containing 5% sheep blood was activated. Because in this medium expression levels of agrA, RNAIII, and hla increased during the stationary phase of growth compared to the exponential phase of growth, the expression of the spa was repressed. However, in the BHI broth, the agr system was a passive system. Because expression of agrA in BHI broth and in stationary phase relative to exponential phase decreased and increasing 3-fold expression of RNAIII can not resulting increasing of expression of hla (Figure 2).
Aside from the growth phase, the expression levels of all genes in cultures containing blood relative to BHI broth alone were increased. The presence of blood in the environment and encounter with blood cells may induce similar conditions to host and the expression profile was similar to those seen with in vivo conditions. It is well known that bacteria with their specific receptors recognize components in the blood, such as heme of hemoglobin, and with an increase in cell density, there is a great increase of virulence genes expression (particularly those affected by the agr regulatory system). Bacteria in the presence of blood (BHIB), which resembles in vivo conditions, compared with BHI broth, will lead agr regulatory system to increase expression of virulence factors. Thus, a 4-fold increase in hla gene expression was observed, which explained the presence of red blood cells, white blood cells, and platelets that could be targeted by this toxin (1, 19). This topic, along with the decrease in the gene expression of spa, indicates the activation of quorum sequencing system by agr in BHI supplemented with 5% sheep blood.
In the current study, in BHI broth, despite an increase in cell density, quorum sequencing system is not activated in a standard laboratory culture media. The probable reason of this subject is that BHI broth is an enriched nutritious media and is ideal for growth of S. aureus and bacteria are not needed for the expression of the agr system and virulence genes. Oogai et al. (2011) showed that levels of expression of effector molecule of the agr locus (RNAIII) in stationary phase increased in TSB broth, yet this increase of regulatory gene of agr system resulted in decreased expression of some virulence genes, similar to aur (encoding aureolysin) and lukS-PV (encoding Panton-Valentine leukocidin) (5).
The impact of the type media culture on the expression of virulent genes has been reviewed and approved by several studies. Malachowa et al. (2011) reported that expression of gamma-hemolysin gene and Pantone Valentin gene in three media cultures BHI, TSB, and CCY is different: Pantone Valentin gene was more expressed in CCY than the other two media cultures and gamma hemolysin gene showed greater expression in BHI. It is interesting that Pantone Valentin gene expression in CCY is much higher in the stationary phase than the exponential phase, while this gene is expressed more in TSB during the exponential phase, and is almost the same in BHI (14).
However, the regulatory role of agr system is not the same in all strains and isolates. The diversity and polymorphism in the agr locus is associated with differences in activity level of strains (20, 21). Therefore, polymorphism in the agr locus may result in fundamental differences in the sequences encoding agrB, agrD, and agrC, and influences the downstream area agr locus and changes the expression levels of agrA. In the current study, it was found that the gene involved in the resistance to methicillin (mecA) was strongly upregulated by agr in the BHI broth containing 5% sheep blood, which is in accordance with the idea that agr impacts methicillin resistance, according to the study of Cheung et al. (22). These results show that agr has a gene regulatory effect on the expression of methicillin resistance gene.
The results indicated that expression of all selected genes increased in colonizing isolates compared to clinical isolates (Figure 4). In other studies, a significant correlation was not found between the presence of virulence genes and invasiveness of S. aureus isolates (23, 24). The origin of the isolate cannot be considered independently of other genetic factors related to the isolate, as these may make a significant contribution to pathogenicity (25).
Results of comparison of gene expression profile in MRSA and MSSA isolates indicated that expression of all selected genes except spa, increased in MSSA isolates compared with MRSA isolates (Figure 5). The acquisition of resistance to antibiotics is decreased toxins expression (26-28). Also, decreased expression of virulence genes was associated with type and size of cassette SCCmec. The MRSA strains carrying the larger cassette SCCmec showed higher energy demand and reduction in virulence factor secretion (23, 29).
5.1. Conclusions
The agr system in the BHI broth, containing 5% sheep blood, was active, as in this medium, expression levels of agrA, RNAIII, and hla increased during the stationary phase of growth compared to the exponential phase of growth, while the expression of the spa was repressed. The presence of blood in the media culture and encounter with blood cells may induce similar conditions to host and the expression profile was similar to those seen with in vivo conditions. Studying the effect of the presence of blood in the medium on the expression of other virulent genes and other regulatory systems, and accurate direct measurement of gene expression levels in-vivo could provide a better understanding of the pathogenesis of S. aureus.