1. Background
Acinetobacter baumannii has emerged as an important agent of hospital-acquired infections, particularly in patients hospitalized in intensive care units (ICUs) as well as surgical wards (1). Ventilator-associated pneumonia (VAP), secondary meningitis, wounds, soft-tissues, bloodstream, and urinary tract infections (UTI) are the most reported infections caused by A. baumannii (2). The risk factors that predispose the development of infection with A. baumannii in hospitalized patients include the prolonged length of hospital stay, number of hospital beds (over 500 beds), major trauma (such as burns and surgeries), and utilization of mechanical ventilators or indwelling catheters. Carbapenems, aminoglycosides, quinolones, and tetracyclines, solely or in combination, are the most antibiotics in the treatment of severe A. baumannii infections (3).
Todays, dramatically increased reporting of multidrug-resistant A. baumannii (MDRAB) outbreaks has attracted special attention to this opportunistic pathogen (4-9). Consequently, emerging extremely-drug resistant (XDR) and pan-drug resistant (PDR) isolates has worsened the problem that highlights the great importance of infection prevention control (10). Wide varieties of antimicrobial resistance mechanisms have been found to contribute to the emergence of MDRAB. The acquisition and transfer of antibiotic resistance genes on plasmids, integrons, transposons, and mutations are the usual mechanisms (11).
Production of β-lactamases is the most prevalent mechanism of β-lactam drug resistance in A. baumannii. Oxacillinases (OXA-23, OXA-24/40, OXA-51, and OXA-58) and metallo-β-lactamases (MBLs), such as IMP, VIM, GIM, and SIM, are enzymes that confer resistance to carbapenems (doripenem, imipenem, and meropenem). The production of the extended spectrum β-lactamases (ESBLs), including blaTEM, blaSHV, and blaCTX, as well as Acinetobacter-derived cephalosporinase (blaADC), plays a critically important role in resistance to cephalosporins in the MDRAB. Furthermore, ISAba1 is one of the most frequently reported insertion sequences in A. baumannii, providing a strong promoter for overexpression of the intrinsic blaADC and some of the oxacillinase genes (OXA-23, OXA-24/40, and OXA-58) (12, 13). Moreover, aminoglycoside-modifying enzymes (AMEs) such as nucleotidyltransferases (aadA1 and aadB), phosphotransferases (aphA1 and aphA6), and acetyltransferase (aacC1) have been detected in A. baumannii. On the other hand, resistance to tetracycline is caused by active efflux pumps (tetA, tetB, and tetH) and ribosomal protection (tetM). In addition, general AdeABC efflux pumps (adeB) contribute to the resistance against β-lactams, tetracyclines, and aminoglycosides (14).
Mashhad, located in the northeast of Iran, is among the holiest cities in the Shia Muslim world. Mashhad is the only major Iranian city that attracts more than 20 million tourists and pilgrims every year, many of whom come to pay homage to the Imam Reza’s shrine (the eighth Shiite Imam). Moreover, political adversity, infirm central government, civil wars, terrorism, poverty, and weak health systems for a long time have affected neighboring countries of Iran and the presence of nearly 4 million foreign nationals from these countries has arrived an additional pressure on the structure of Iran’s health care, especially in Mashhad. Therefore, the transmission of infectious diseases from these countries has changed epidemiological features of these diseases in Iran. Thus, it is important to investigate the molecular epidemiology of infectious diseases including nosocomial infections in this city. Likewise, it should be noted that several A. baumannii outbreaks have been documented in other regions of Iran (15-19). In the previously published report from the Ghaem referral Hospital in Mashhad, we discussed the molecular epidemiology and resistance profile of A. baumannii isolates in detail (5). Thereafter, infection prevention interventions were regarded to eradicate resistant isolates.
2. Objectives
This report includes a six-month period after the previously mentioned study to evaluate the infection-control measures, with focusing on the molecular resistance profile and the clonal relatedness of strains that emerged after the outbreak.
3. Methods
3.1. Ethics Statement
The study protocol was reviewed and approved by the Biomedical Research Ethics Committee of Mashhad University of Medical Sciences (IR.MUMS.REC.89427).
3.2. Hospital Setting and Bacterial Isolates
This prospective cross-sectional study was performed at Ghaem teaching Hospital (Mashhad, northeast-Iran). Ghaem center is a 1,000-bed referral Hospital affiliated to Mashhad University of Medical Sciences (MUMS). In the present study, all A. baumannii strains were collected from clinical cultures. In total, 39 consecutive non-repetitive isolates were obtained from five distinct wards, including neonatal intensive care unit (NICU), ICU, surgery (SUR), internal medicine (IM), and pediatrics (PED) from July to the end of December 2013, after a reported outbreak in our center (5). All A. baumannii isolates were identified by biochemical standard tests (oxidase, oxidative-fermentative (OF) glucose, nitrate reduction, sugars fermentation on TSI, motility on SIM, citrate utilization on Simon’s Citrate agar, growth at 42°C and on MacConkey agar) (20). Moreover, the final confirmation of A. baumannii isolates was performed by PCR of blaOXA-51-like gene and multiplex of gyrB (21).
3.3. Infection Prevention Control (Ipc) Interventions
First, to realize the importance of the issue, a hand hygiene campaign was held. Critical instructions of control infection were published in pamphlets and banners and placed on public view. Special emphasis was placed on hands washing. Furthermore, commercial hand washing solutions were installed on corridors for public access. Multifaceted IPC interventions, including face-to-face interviews with the lead infection control professionals and antimicrobial stewardship programs (ASP), were provided for hospital personnel by the control infection team. Furthermore, wearing gloves and gowns before and after patient care, contact isolation of patients who were colonized or infected by MDRAB, monthly sampling of ICU instruments (shower, laryngoscopes, oxygen flow meters, and other medical equipment) were considered by all healthcare staff. In addition, surfaces in the ICUs were disinfected daily with an aldehyde-containing disinfectant. If needed, the disinfectant was changed from one brand to another. Everywhere MDRAB and other MDR Gram-negative infections were suspected, ampicillin/sulbactam or a carbapenem with an aminoglycoside was used. Colistin was only prescribed for patients in whom carbapenems-resistant A. baumannii (CRAB) was confirmed by susceptibility testing. The protocols were reviewed every month to eradicate probable new outbreaks.
3.4. Antimicrobial Susceptibility Testing
Initially, the antimicrobial susceptibility of each isolate was determined by Kirby-Bauer disk diffusion method on Mueller-Hinton agar (formula adjusted; HiMedia, India) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (22). Therefore, 15 antibacterial agents were tested, including piperacillin (PC), ceftazidime (CA), cefotaxime (CTX), ceftriaxone (CI), cefepime (CPM), imipenem (I), meropenem (MR), gentamicin (G), tobramycin (TB), amikacin (AK), tetracycline (T), doxycycline (DO), ciprofloxacin (CF), and co-trimoxazole (Co) according to the manufacturer’s instructions (HiMedia Diagnostics, India). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 reference strains were used for quality control in the antimicrobial susceptibility testing. Moreover, MICs of all the isolates were determined by E-test strips for four antibacterial agents (selected for MDR detection), including imipenem (IMI), colistin (CS), ceftazidime (CAZ), and ciprofloxacin (CIP) (Liofilchem, Italy). E-test results were interpreted using the CLSI standards criteria for Acinetobacter spp. (22).
3.5. Genomic DNA Preparations
In order to obtain genomic DNA extraction, initially, 39 A. baumannii strains were grown overnight at 37 °C on Mueller-Hinton agar plates until the recovery of fresh culture from each isolate. Then, a loopful of each pure bacterium was suspended in 300 μL of sterile distilled water and boiled for 10 min. After 5 min of centrifugation (10.000 g), the supernatant was kept at -20 °C as DNA template for PCR amplification (23).
3.6. REP-PCR and International Clonal Lineage Typing
Typing of isolates was performed by PCR amplification of repetitive extragenic palindromic elements (REP-PCR), as described previously (24). PCR amplification was performed in a reaction mixture containing REP1 and REP2 primers (1µM), dNTPs mixture (0.2 mM), MgCl2 (3 mM), 1 unit Taq DNA polymerase (Thermo Scientific, USA), 1x PCR buffer, and bacterial DNA template (2 µL) in a final volume of 50 µL. The amplification reaction was carried out by using a thermal cycler (Mastercycler Eppendorf, Germany) with an initial denaturation at 94°C for 10 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 3 min, and a final extension at 72°C for 16 min. The products were separated by electrophoresis on 1.2% agarose gel (Invitrogen, USA) after ethidium bromide staining. Pictures were analyzed by GelJ software (v. 1.3) considering dice tolerance 1.0 and UPGMA method to construct a dendrogram (25). International clonal lineages were determined by multiplex PCR for two groups of genes, consisting of csuE, OXA-66/69, and ompA, as previously described (26).
3.7. PCR Amplification of Resistance Genes
In the present study, we investigated four important groups of MDRAB genes, including efflux pumps (tetA, tetB, and adeB), aminoglycoside-modifying enzymes (aphA1, aphA6, aacC1, aadB, and aadA1), oxacillinases (blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-58), ESBLs (blaTEM and blaADC), metallo-β-lactamases (blaIMP, blaVIM), and ISAba1. A combination of forward primer of ISAba1 and reverse primers of blaOXA-like families and blaADC were used to find the upstream location of IS element in the mentioned genes (27). The list of primer sequences and product length is shown in Table 1. Unfortunately, we were not able to prepare resistant reference strains and control target genes; thus, we overcame this pitfall by alternative access pathways. If any PCR reaction was negative for one sample, the PCR procedure was repeated twice while first positive samples were sequenced (Macrogen, Seoul, Korea; http://www.macrogen.com) for confirmation. Thereafter, we used the confirmed sequence as a positive control. Finally, all the amplified products were analyzed by electrophoresis on 1.5% agarose gel in 0.5x TBE buffer. Agarose gel was stained by 0.5x TBE buffer containing 0.5 μg/mL ethidium bromide, with an exception of REP-PCR products that were separated on 1.2% agarose gel, and viewed by UV Geldoc apparatus (uvitec, UK).
| Target genes | Forward Sequences (5’ → 3’) | Reveres Sequence (5’ → 3’) | Annealing, °C | Amplicon Size, bp | References |
|---|---|---|---|---|---|
| Tetracycline resistance genes (efflux genes) | |||||
| tetA | GGCGGTCTTCTTCATCATGCA | CGAAGGCAAGCAGGATGTAGC | 54 | 242 | This study |
| tetB | TACGTGAATTTATTGCTTCGG | ATACAGCATCCAAAGCGCAC | 59 | 206 | (28) |
| adeB | GGATTATGGCGACAGAAGGA | AATACTGCCGCCAATACCAG | 52 | 105 | (29) |
| Aminoglycoside-modifying enzyme genes | |||||
| aphA1 | GCGTTGCCAATGATGTTACAG | CGAGCATCAAATGAAACTGC | 51 | 623 | (30) |
| aphA6 | ATGGAATTGCCCAATATTATTC | TCAATTCAATTCATCAAGTTTTA | 55 | 780 | (31) |
| aacC1 | TTAGGTGGCGGTACTTGGGTC | ATGGGCATCATTCGCACATGTAGG | 64 | 421 | (31) |
| aadB | ATGGACACAACGCAGGTCGC | TTAGGCCGCATATCGCGACC | 70 | 495 | (31) |
| aadA1 | ATGAGGGAAGCGGTGATCG | TTATTTGCCGACTACCTTGGTG | 64 | 792 | (31) |
| blaOXA-like carbapenemase genes | |||||
| blaOXA-23-like | TGTTGAATGCCCTGATCGGA | TACGTCGCGCAAGTTCCTGA | 55 | 167 | This study |
| blaOXA-24-like | GGTTAGTTGGCCCCCTTAAA | AGTTGAGCGAAAAGGGGATT | 52 | 246 | (32) |
| blaOXA-51-like | CGTGCTTCGACCGAGTATGTAC | GACTTGGGTACCGATATCTGCA | 55 | 276 | This study |
| blaOXA-58-like | AAGTATTGGGGCTTGTGCTG | CCCCTCTGCGCTCTACATAC | 52 | 599 | (32) |
| Metallo-β-lactamases genes and ISAba1 | |||||
| blaIMP | GGAATAGAGTGGCTTAAYTCTC | CCAAACYACTASGTTATCT | 52 | 188 | (33) |
| blaVIM | GATGGTGTTTGGTCGCAT | CGAATGCGCAGCACCAG | 52 | 390 | (33) |
| blaADC | ATGCGATTTAAAAAAATTTCTTGT | TGGAATACGTTTATTGGTTAACATGA | 50 | 1081 | (31) |
| blaTEM | GCACGAGTGGGTTACATCGA | GGTCCTCCGATCGTTGTCAG | 65 | 310 | (31) |
| ISAba1 | CACGAATGCAGAAGTTG | CGACGAATACTATGACAC | 56 | 549 | (27) |
4. Results
4.1. Bacterial Isolates and IPC Interventions Outcomes
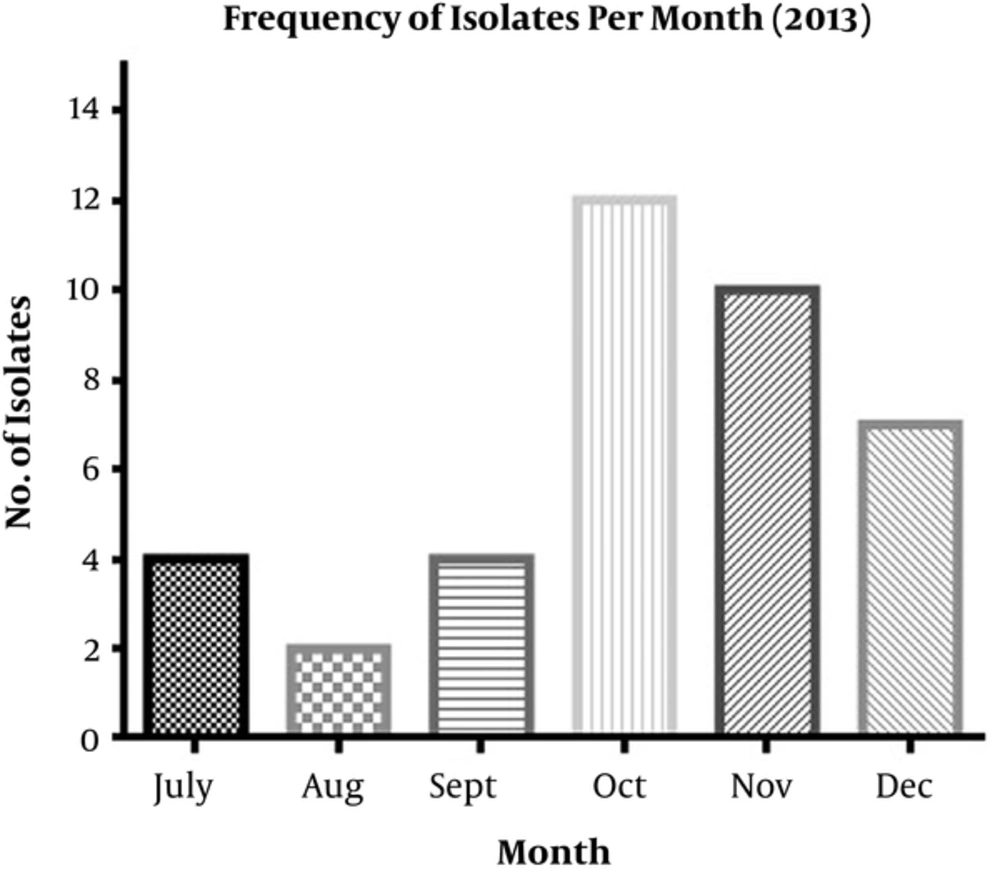
In total, 39 A. baumannii isolates were obtained from hospitalized patients, including 20 (51%) females and 19 (49%) males aged 1 to 70 years. The isolates were recovered from respiratory sites (n = 16; 41%), blood (n = 14; 36.8 %), wound (n = 2; 5.2%), CSF (n = 2; 5.2%), urine (n = 1; 2.5%), eye (n = 1; 2.5%), and blister (n = 1; 2.5%). Moreover, two isolates (5.2%) were recovered from health-care associated devices (HAD). Of the 39 A. baumannii isolates collected, 17 (43.5%), 12 (31%), seven (18%), two (5%), and one (2.5%) were isolated from the ICU, NICU, SUR, IM, and PED wards, respectively. Both isolates that were originated from HADs were recovered in NICU. The numbers of isolates per month and bacteria isolation date are illustrated in Figure 1. The number of isolates gradually decreased from July to September 2013 but dramatically increased in October. According to the data presented in Figure 1, it could be concluded that the IPC was successful in the first three months of the program, after which it was neglected gradually.
4.2. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility testing revealed that the most effective antibacterial agent was colistin, for which all isolates were susceptible to colistin (Table 2). Moreover, 51% of the isolates were sensitive to doxycycline. All isolates were completely resistant to tested β-lactam antibiotics, including third-generation cephalosporins, carbapenems, and piperacillin. These results show that all isolates were MDRAB. The MIC of the isolates by E-test showed a high rate of resistance to imipenem, ceftazidime, and ciprofloxacin; however, colistin stood as the only effective antibiotic (MIC ≤ 2 µg/L) as shown by disk diffusion method. The E-test results are summarized in Table 2.
| Isolate No. | IC | REP-Type | E-Test | Source | Ward | tetA | tetB | adeB | aphA1 | aphA6 | aacC1 | aadB | aadA1 | blaIMP | blaVIM | blaADC | blaTEM | OXA-23 | OXA-24 | OXA-51 | OXA-58 | ISAba1 | ISAba1/ OXA | ISAba1 /blaADC | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | IMI | CS | CIP | ||||||||||||||||||||||||
| 1 | I | E | > 256 | > 32 | 2 | > 32 | W.C | SUR | - | + | + | + | - | - | + | + | - | + | + | + | + | + | + | - | + | 23 | + |
| 2 | I | C2 | 32 | > 32 | 2 | > 32 | T.A | ICU | - | + | + | + | + | - | + | + | - | + | + | + | + | + | + | - | + | 23 | + |
| 3 | II | E | > 256 | > 32 | 2 | > 32 | B.C | NICU | - | + | + | + | - | + | + | + | - | + | + | + | + | + | + | - | + | 23 | + |
| 4 | I | C2 | > 256 | > 32 | 2 | > 32 | T.A | ICU | - | + | + | + | + | - | + | + | - | + | + | + | + | + | + | - | + | 23 | _ |
| 5 | II | D | > 256 | > 32 | 2 | > 32 | B.C | NICU | - | + | + | + | + | - | + | + | - | + | + | + | + | + | + | - | + | 23 | _ |
| 6 | I | C2 | 96 | > 32 | 1.5 | > 32 | T.A | ICU | - | + | + | - | + | + | + | + | - | + | + | + | + | + | + | - | + | 23 | _ |
| 7 | II | D | > 256 | > 32 | 1.5 | > 32 | B.C | NICU | - | + | + | - | + | + | + | + | - | - | + | + | + | - | + | - | + | 23 | + |
| 8 | I | E | > 256 | > 32 | 1 | > 32 | B.C | NICU | - | + | + | + | + | + | + | + | - | + | + | + | + | + | + | - | + | 23 | + |
| 9 | I | C2 | > 256 | > 32 | 2 | > 32 | HAD | NICU | - | + | + | - | + | + | + | + | - | - | + | + | + | + | + | - | + | 23 | _ |
| 10 | II | D | > 256 | > 32 | 2 | > 32 | B.C | NICU | - | + | + | - | + | + | + | + | - | - | + | + | + | + | + | - | + | 23 | _ |
| 11 | II | D | > 256 | > 32 | 2 | > 32 | B.C | NICU | - | + | + | + | - | - | + | + | - | + | + | + | + | + | + | - | + | 23 | + |
| 12 | II | D | > 256 | > 32 | 1.5 | > 32 | B.C | SUR | - | + | + | + | - | - | + | + | - | - | - | + | + | + | + | - | + | 23 | _ |
| 13 | I | C2 | > 256 | > 32 | 1.5 | > 32 | B.C | ICU | - | + | + | - | + | - | + | + | - | - | + | + | + | - | + | - | + | 23 | _ |
| 14 | UN | C1 | > 256 | > 32 | 2 | > 32 | B.C | IM | - | + | + | - | + | - | + | + | - | + | + | + | + | + | + | - | + | 23 | _ |
| 15 | I | E | > 256 | > 32 | 2 | > 32 | HAD | NICU | - | + | + | + | - | - | + | + | - | - | + | + | + | - | + | - | + | 23 | _ |
| 16 | I | E | > 256 | > 32 | 2 | > 32 | B.C | NICU | - | + | + | + | - | - | + | + | - | - | + | + | + | - | + | - | + | 23 | _ |
| 17 | I | E | > 256 | > 32 | 1 | > 32 | B.C | NICU | - | + | + | - | + | - | + | + | + | - | + | + | + | + | + | - | + | 23 | _ |
| 18 | I | E | > 256 | > 32 | 2 | > 32 | B.C | NICU | - | + | + | - | + | + | + | - | - | - | + | + | + | - | + | - | + | 23 | _ |
| 19 | I | C2 | > 256 | > 32 | 2 | > 32 | T.A | ICU | - | + | + | + | + | - | + | + | - | + | + | + | + | + | + | - | + | 23 | _ |
| 20 | UN | C2 | > 256 | > 32 | 2 | > 32 | E.D | NICU | - | + | + | + | - | + | + | + | - | + | - | + | + | - | + | - | + | 23 | _ |
| 21 | I | C2 | > 256 | > 32 | 1.5 | > 32 | T.A | ICU | - | + | + | - | + | - | + | + | - | - | + | + | + | + | + | - | + | 23 | + |
| 22 | UN | C2 | > 256 | > 32 | 1 | 0.064 | CSF | ICU | - | + | - | - | + | - | + | + | - | + | + | + | + | + | + | - | + | 23 | + |
| 23 | UN | C2 | > 256 | > 32 | 2 | > 32 | W.C | IM | - | + | + | - | + | - | + | + | - | - | + | + | + | - | + | - | + | 23 | + |
| 24 | UN | C2 | > 256 | > 32 | 2 | > 32 | T.A | ICU | - | + | - | - | - | - | + | + | - | - | - | + | + | - | + | - | + | 23 | _ |
| 25 | I | C2 | > 256 | > 32 | 1 | > 32 | T.A | ICU | - | + | + | - | + | - | + | + | - | + | + | + | + | - | + | - | + | 23 | _ |
| 26 | II | D | > 256 | > 32 | 1 | > 32 | W.C | SUR | - | + | + | - | + | + | + | - | - | + | - | + | + | + | + | - | + | 23 | + |
| 27 | I | C2 | > 256 | > 32 | 1 | > 32 | T.A | ICU | - | + | + | + | + | - | + | + | - | + | + | + | + | + | + | - | + | 23 | + |
| 28 | I | C2 | > 256 | > 32 | 1 | > 32 | T.A | ICU | - | + | + | - | - | + | + | + | - | + | + | + | + | - | + | - | + | 23 | _ |
| 29 | I | C2 | > 256 | > 32 | 1 | > 32 | T.A | ICU | - | + | + | - | + | - | + | + | - | + | + | + | + | + | + | - | + | 23 | _ |
| 30 | I | E | > 256 | > 32 | 1.5 | > 32 | T.A | PED | - | + | + | - | + | + | + | + | - | + | + | + | + | + | + | - | + | 23 | _ |
| 31 | I | C2 | > 256 | > 32 | 2 | > 32 | T.A | ICU | - | + | + | + | - | - | + | + | - | - | + | + | + | + | + | - | + | 23 | _ |
| 32 | UN | B | > 256 | > 32 | 1 | > 32 | T.A | SUR | - | + | + | - | + | - | + | + | - | + | - | + | + | + | + | - | + | 23 | _ |
| 33 | I | C2 | > 256 | > 32 | 1.5 | > 32 | CSF | ICU | - | + | + | - | - | - | + | + | - | - | - | + | + | + | + | - | + | 23 | _ |
| 34 | I | C1 | > 256 | > 32 | 2 | > 32 | T.A | SUR | - | + | + | + | + | - | + | + | - | + | + | + | + | - | + | - | + | 23 | + |
| 35 | I | C2 | > 256 | > 32 | 2 | > 32 | T.A | SUR | - | + | + | + | + | + | + | + | - | + | + | + | + | + | + | - | + | 23 | + |
| 36 | UN | A | 96 | > 32 | 1 | > 32 | U.C | SUR | - | + | + | + | + | + | + | + | - | + | + | + | + | + | + | - | + | 23 | _ |
| 37 | UN | B | > 256 | > 32 | 0.5 | > 32 | B.C | NICU | - | + | + | + | + | + | + | - | - | + | + | + | + | + | + | - | + | 23 | + |
| 38 | II | D | > 256 | > 32 | 1 | > 32 | B.C | NICU | - | + | + | + | + | + | + | + | - | + | - | + | + | + | + | - | + | 23 | + |
| 39 | II | D | > 256 | > 32 | 1 | > 32 | T.A | NICU | - | + | + | + | + | + | + | + | + | - | + | + | + | - | + | - | + | 23 | - |
| R, % | 100 | 100 | 0 | 100 | |||||||||||||||||||||||
| Total, % | 0 | 39 (100) | 37 (95) | 21 (54) | 28 (72) | 16 (41) | 39 (100) | 36 (92) | 2 (5) | 24 (61) | 32 (82) | 39 (100) | 39 (100) | 27 (70) | 39 (100) | 0 | 39 (100) | 39 (100) | 15 (38.4) | ||||||||
Abbreviations: B.C, blood culture; CAZ, ceftazidime; CIP, ciprofloxacin; CS, colistin; CSF, cerebral spinal fluid; E.D, eye discharge; HAD, health-care associated device; ICU, intensive care unit; IM, internal medicine; IMI, imipenem; NICU, neonatal intensive care unit; PED, pediatrics; SUR, surgery; T.A, tracheal aspirates; U.C, urine culture; W.C, wound culture.
4.3. Resistance Gene Determinants and Clonality Analysis
This study evaluated a variety of resistance genes in A. baumannii. OXA-51, OXA-23, and blaTEM were detected in all isolates while OXA-58 was not detected in any isolates. Other β-lactamases such as blaADC, blaVIM, blaIMP, and OXA-24 were found in 82% (31/39), 61% (24/39), 5% (2/39), and 70% (27/39) of the isolates, respectively. Alongside resistance genes, ISAba1, as an intrinsic insertion sequence to A. baumannii, was characterized in all isolates. Upstream-located ISAba1 in β-lactamases was detected along with 39 (100%) OXA-23 and 15 (38%) blaADC producer isolates.
Efflux pumps, tetB and adeB, were widely distributed in isolates so that all of them produced these two types of efflux pumps. In the present study, adeB and tetB were detected in 95% (37/39) and 100% (39/39) of the A. baumannii isolates. Interestingly, tetA was not detected in any of the isolates. Aminoglycoside-modifying enzymes, including aphA1, aphA6, aacC1, aadB, and aadA1, were found to be extensively disseminated in all isolates, of which aadB was the most distributed; hence, all isolates presented this gene. Other genes such as aadA1, aphA6, aphA1, and aacC1 were detected in 92% (36/39), 72% (28/39), 54% (21/39), and 41% (16/39) of the isolates. Moreover, 37 (95%) isolates possessed more than three AME genes (Table 2).
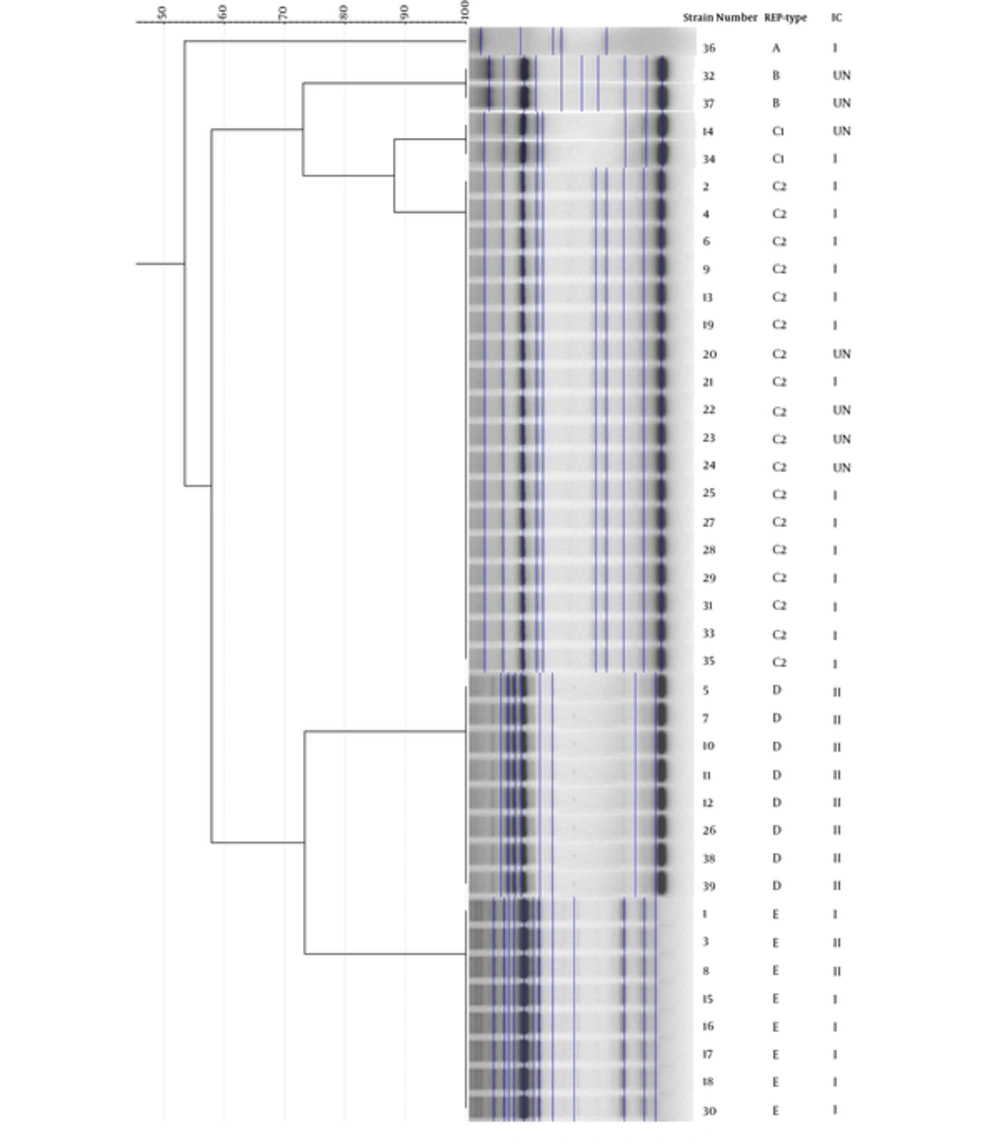
To determine the clonal relationship, all isolates were analyzed via REP-PCR method (Figure 2). According to the REP-PCR band pattern, 20 (51 %) strains belonged to cluster C. Disseminations of clones were not limited to a specific ward; however, most of the strains that isolated from ICU and NICU belonged to clusters D and E, respectively (Table 2). PCR-based international clonal lineage detection determined that 22 (56.5%) and nine (23%) strains were from IC1 and IC2, respectively. The lineages of eight (20.5%) strains were unknown. The largest cluster (C) belonged to IC1, which mostly was isolated from ICUs (Table 2).
5. Discussion
The acquisition of antimicrobial resistance determinants increases the ability of A. baumannii strains to survive in the hospital setting (34). In the present study, most of the isolates were recovered from tracheal aspirates and blood. It was not surprising since A. baumannii mostly encounters nosocomial infections. Therefore, infection control should be regarded as the core priority in hospitals. At the start point of the IPC program in our center (July 2013), healthcare personnel was alerted to the issue, and the consequence of program was successful since A. baumannii colonization and infection reduced. Nevertheless, it continued only for three months where A. baumannii isolation increased. Since a new clone did not emerge in the second trimester of the study, the weak points of the study must be cleared. It should be stressed that infection control is somehow changing cultural behaviors rather than following written roles (35). However, it is worth noting that based on the molecular approach, we could realize the efficacy of IPC intervention and epidemiological context.
In the present study, colistin showed an excellent activity against MDRAB strains. Moreover, 51% of the A. baumannii isolates were found to be susceptible to doxycycline. Other agents, including carbapenems and ceftazidime, showed high resistance rates. The high rate of resistance to antimicrobials, especially those belonging to β-lactam and aminoglycosides, might be related to the bias in a wide prescription of these antibiotics (Table 2). It should be noted that tetracycline and doxycycline were not prescribed for A. baumannii infections in our center while the isolates were moderately resistant to these agents. It can be deduced that resistance to antibacterial agents did not depend on their prescription, and resistance genes had been acquired without antibiotic pressure but probably through resistance plasmids (36).
Since carbapenems are the last resort of antibiotics for refractory isolates, resistance to this group has raised a great concern. Carbapenem resistance has been attributed to the overproduction of oxacillinases by ISAba1 located upstream and to a lesser extent, MBLs production (37). In recent years, OXA carbapenemase-producing A. baumannii has been increasingly reported from Asian countries and MBL genes such as blaIMP and blaVIM have been reported rather sporadically (38). In Iran, few studies reported MBLs as the causative of carbapenem production (39, 40). However, our results revealed that 61% of the strains harbored blaVIM. In contrast to our results, those of Salimizand et al. did not detect MBLs at the other Hospital near to our center in 2012 (8). Studies by Sohrabi et al. and Salimizand et al. confirmed the role of adjacent IS to OXA-families (8, 41). In this study, OXA-23-like and OXA-24-like were the most prevalent oxacillinase genes, while OXA-58-like was not detected among the MDRAB strains. In the present study, we found that the coexistence of OXA-23/OXA-24 was more common among the majority of MDR strains (70%). We also identified that all isolates of A. baumannii harbored the ISAba1 upstream of OXA-23, which has proven to confer carbapenems resistance. The location of ISAba1 upstream of OXA-23 is responsible for its overexpression (8). The extended distribution of OXA-23 adjacent of transposable elements (ISAba1) in Iranian A. baumannii isolates indicates a plasmid-borne transfer among the species. Therefore, more studies are required to find the reason for carbapenem resistance by MBLs or overproduction of oxacillinases. Obviously, real-time PCR of interesting genes will help understand this issue. Moreover, our results revealed that the presence of blaTEM (100%) and blaADC (82%) in most clinical isolates could be responsible for cephalosporin resistance.
With regard to the previous studies in this region, the most prevalent AMEs were aac1 and aphA6 (42-44) while in our study, aadB (100%) was the most detected AMEs (Table 2). It should be noted that most isolates (37/39) carried more than three types of AMEs. The antibiotic susceptibility testing showed that all isolates were fully resistant to at least one of gentamicin, tobramycin, or amikacin and at least one resistance gene was detected in each isolate. Apart from detecting AMEs in A. baumannii, studying other factors such as alteration of the ribosomal binding site and reduced expression of efflux pumps, which are committed in aminoglycoside non-susceptibility, can make a better perspective of resistance determinants in A. baumannii (45).
Efflux pumps have an undeniable effect in emerging refractory bacteria as much as resistance genes do, as observed in A. baumannii (46). Considering the widespread efflux pumps in A. baumannii, the major facilitator superfamily (MFS) and the resistance-nodulation-division (RND) families, we assessed the presence of adeB (from RND), tetA, and tetB (from MFS in our isolates) (Table 2). On the other hand, tetB and adeB were found to be in 100% and 95% of the isolates, respectively, and tetA was not detected in any isolate. These results were surprisingly different from other centers in this region and others elsewhere (24, 44, 47, 48). Hereafter, adeB, tetB, and probably other tetracycline resistance determinants, such as tetH, tetM, and tetX, were the dominant reason for resistance to tetracyclines. More work should be performed to find the exact resistance profile of collection.
Determining the genetic determinants of MDRAB in medical centers may help eradicate nosocomial infections through tracking similar isolates in hospitalized patients. REP-PCR is a simple, feasible, and low-cost method compared to other methods. Thus, this technique has been verified to have acceptable discrimination and reproducibility for studying outbreaks of A. baumannii (24, 49). Since clonally related strains were isolated from different wards, it suggests cross-transmission of strains. Furthermore, REP-PCR generated band patterns of strains in this study were not mainly differed from the patterns observed in the previously reported outbreak in our center (data not shown). According to the evidence presented in Table 2, it was interesting that the majority of strains (87.1%) belonged to two international clonal lineages, I and II. It confirms that these two clones are refractory to antibacterials and circulate among patients in different wards of our center.
In terms of clonality relatedness of isolates, there are other techniques with reliable discriminatory power. Among them, multi-locus sequence typing (MLST) with a global database (www.pubmlst.org) is well accepted by investigators from different parts of the world. This sequence-based typing system nominated isolates as sequence type (ST) according to their sequence of housekeeping genes (50). One of the limitations of the present study was the REP-PCR typing procedure that had intra-center reproducibility (51). Several reports from Iran that have used MLST technique showed that the most prevalent A. baumannii ST in Iran was ST2 that has been in circulation in this country (18, 52-55). Since most of the IC2 strains belonged to ST2, it was probable that our center is colonized with this ST. Further studies are needed to find and clear the prevalent STs in this center.
6. Conclusions
The results revealed that the clonal distribution of A. baumannii in different wards of Ghaem Hospital. The presence of numerous resistance genes led to a high level of resistance to most of the antibiotics. Moreover, the emergence of epidemic MDR A. baumannii clones in the northeast of Iran indicates the necessity of a screening program for patients after hospitalization abroad. Stringent infection control procedures to prevent further dissemination are urgent. However, as a serious concern, a further study is essential to evaluate the effectiveness of infection-control measures.