1. Background
Colorectal cancer (CRC) is a highly prevalent malignancy worldwide, ranking as the third most common cancer after breast and lung cancers, with a higher incidence observed in men. Various risk factors, including diet, lifestyle, obesity, heredity, and inflammation, contribute to CRC development (1, 2). Microbiota dysbiosis is a common feature in the pathogenesis of certain cancers (3, 4). Notably, an increased presence of Fusobacterium nucleatum in CRC patients compared to healthy individuals has emerged as a significant marker for CRC diagnosis (5). Studies have shown that F. nucleatum promotes CRC progression by activating signaling pathways such as E-cadherin/β-catenin and TLR4/MyD88, and modulating autophagy and immune responses (6). Moreover, F. nucleatum is implicated in various molecular events, including microsatellite instability (MSI), CpG island methylator phenotype (CIMP), and tumorigenic mutations in genes such as TP53, BRAF, CHD7, and CHD8, which may contribute to tumor initiation and progression (7).
Key factors in F. nucleatum pathogenesis include Fusobacterium adhesin A (FadA), which facilitates cell attachment, lipopolysaccharides (LPS) that activate proinflammatory genes and promote CRC progression through the release of inflammatory cytokines, and Fusobacterium autotransporter protein 2 (Fap2), another virulence factor enhancing CRC receptor cell binding. Additionally, F. nucleatum has been shown to induce epigenetic alterations in CRC (8-10). Reactive oxygen species (ROS) and inflammatory cytokines produced by F. nucleatum contribute to the silencing of the mismatch repair protein (mlh1) gene, resulting in MSI tumors (11). Dysfunction in this repair system, often arising from epigenetic silencing, leads to an accumulation of mutations in the genome (12). Given the variations in F. nucleatum prevalence across different ethnicities and geographical regions, MMR status may differ accordingly (13). Understanding the interplay between F. nucleatum and MMR genes may provide valuable insights into the molecular mechanisms of CRC development.
2. Objectives
In this study, we aim to investigate the effects of F. nucleatum on the relative expression of mlh1, msh2, and msh6 genes in Iranian CRC patients.
3. Methods
3.1. Study Design and Sample Collection
A case-control study design was employed to compare the relative prevalence of F. nucleatum and the expression of mlh1, msh2, and msh6 genes in CRC patients versus a control group. This research was conducted at the Microbiology Department of Alborz University of Medical Sciences from January 2022 to February 2023. Biopsy specimens were collected from 40 CRC patients and 20 individuals with suspected CRC who underwent colonoscopies. The specimens were obtained by a gastroenterologist from the colon and rectum. Eligibility for the study required CRC patients to be aged 18 or older and to provide written informed consent. Patients who had received systemic chemotherapy were excluded. Specimens were transported to the Microbiology Department in Transystem tubes containing normal saline and RNA-later and stored at -20°C until further processing.
3.2. DNA and RNA Extraction, and cDNA Synthesis
DNA extraction from the tissue samples was carried out using an extraction kit (Iran ROJE Co.), following the manufacturer's specifications. Bacterial cells were concentrated by centrifugation, and their DNA was extracted. The concentration and purity of the DNA samples were assessed using a spectrophotometer (NanoDrop 2000). RNA was extracted from the specimens using an RNA extraction kit (Iran ROJE Co.) per the manufacturer's instructions. The quality of the extracted RNA was also evaluated using a spectrophotometer (NanoDrop 2000). Complementary DNA was then synthesized from the RNA samples using a cDNA synthesis kit (Iran ROJE Co.).
3.3. Fusobacterium nucleatum Detection
The presence of F. nucleatum in the samples was determined using real-time PCR targeting the specific 16srRNA gene of F. nucleatum with the following program: Initial denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 20 seconds, 56°C for 30 seconds, and 72°C for 30 seconds. Real-time PCR using universal 16srRNA primers assessed the relative abundance of F. nucleatum in CRC and control groups.
3.4. Expression Analysis of mlh1, msh2, and msh6 Genes
Real-time PCR was conducted to measure the relative expression levels of the mlh1, msh2, and msh6 genes in both CRC and control groups. The expression levels were normalized against a reference gene (gapdh) for each sample. Real-time PCR was carried out using the Applied Biosystems 7900 system with SYBR® Select Master Mix (Bioneer, Korea) in 20 µL reactions. The cycle conditions for the mlh1, msh2, and msh6 genes included an initial denaturation step of 10 minutes at 95°C, followed by 40 cycles of 20 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C. All reactions were performed in triplicate.
3.5. Statistical Analysis
The presence, relative frequency of F. nucleatum, and the relative expression of the mlh1, msh2, and msh6 genes were analyzed in both the control group (n = 20) and the CRC group (n = 40) based on biopsy samples. The relative expression of each MMR gene compared to gapdh RNA was determined using the formula 2(-ΔCt), where ΔCt represents Ct (Target) - Ct (Reference). The fold change of target gene expression was calculated using the below formula:
Data were analyzed using SPSS version 21 and GraphPad PRISM software version 8. Quantitative data were summarized as mean ± standard deviation. The normality of the data was assessed, and, if appropriate, non-parametric analysis of variance (ANOVA) with a significance level (P-value < 0.05) was applied.
4. Results
4.1. Patients and Samples
The cancer group comprised 52% women and 48% men, with median ages of 55 and 65 years for women and men, respectively. The control group consisted of 45% women and 55% men, with the highest age ranges being 30 - 40 for women and 30 - 50 for men. Colonoscopy was performed based on signs and symptoms such as anemia (34%), abdominal pain (31%), blood in the stool (19%), and rectal bleeding (16%). The tissue samples analyzed included adenocarcinoma (87%) and adenoma (13%), taken from both proximal and distal sections of the intestine. A detailed description of the cancer samples is provided in Table 1.
| Patient ID | Tumor | ||||
|---|---|---|---|---|---|
| Age, y | Sex | Location | Size | Morphology | |
| C01 | 42 | F | Ascending colon | 0.3X0.2X0.1 | Adenocarcinoma |
| C02 | 59 | M | Hepatic flexure | 0.5X0.4X0.2 | Adenocarcinoma |
| C03 | 72 | M | Rectum | 1.5X1X0.3 | Adenocarcinoma |
| C04 | 82 | F | Sigmoid colon | 1.5X1X0.7 | Adenocarcinoma |
| C05 | 69 | F | Sigmoid colon | 0.5X0.4X0.2 | Adenocarcinoma |
| C06 | 51 | M | Descending colon | 0.3X0.2X0.1 | Adenocarcinoma |
| C07 | 49 | M | Sigmoid colon | 0.3X0.2X0.1 | Adenocarcinoma |
| C08 | 78 | M | Cecum | 0.3X0.2X0.1 | Adenocarcinoma |
| C09 | 68 | M | Cecum | 0.5X0.3X0.2 | Adenocarcinoma |
| C10 | 48 | F | Rectum | 0.8X0.6X0.2 | Adenoma |
| C11 | 76 | F | Cecum | 1X1X0.3 | Adenocarcinoma |
| C12 | 27 | M | Ascending colon | 0.6X0.4X0.2 | Adenocarcinoma |
| C13 | 51 | F | Rectum | 1X0.7X0.3 | Adenocarcinoma |
| C14 | 84 | M | Rectum | 0.3X0.2X0.1 | Adenoma |
| C15 | 70 | F | Rectum | 0.3X0.2X0.1 | Adenocarcinoma |
| C16 | 76 | F | Hepatic flexure | 0.3X0.2X0.1 | Adenocarcinoma |
| C17 | 56 | F | Sigmoid colon | 0.5X0.3X0.2 | Adenocarcinoma |
| C18 | 65 | M | Ascending colon | 0.5X0.3X0.2 | Adenocarcinoma |
| C19 | 51 | M | Sigmoid colon | 1X0.7X0.3 | Adenocarcinoma |
| C20 | 49 | M | Sigmoid colon | 0.3X0.2X0.1 | Adenocarcinoma |
| C21 | 63 | F | Sigmoid colon | 1X0.8X0.2 | Adenoma |
| C22 | 58 | M | Sigmoid colon | 0.9X0.7X0.3 | Adenocarcinoma |
| C23 | 64 | M | Descending colon | 0.3X0.2X0.1 | Adenocarcinoma |
| C24 | 52 | M | Rectum | 1X0.9X0.2 | Adenocarcinoma |
| C25 | 58 | F | Ascending colon | 0.6X0.2X0.2 | Adenoma |
| C26 | 45 | M | Descending colon | 0.7X0.5X0.2 | Adenocarcinoma |
| C27 | 56 | F | Rectum | 0.3X0.2X0.1 | Adenocarcinoma |
| C28 | 86 | M | Rectum | 0.3X0.2X0.1 | High grade glandular dysplasia |
| C29 | 73 | M | Rectum | 0.3X0.2X0.1 | Adenocarcinoma |
| C30 | 59 | F | Rectum | 0.3X0.2X0.1 | Adenocarcinoma |
| C31 | 63 | F | Rectum | 0.3X0.2X0.1 | Adenocarcinoma |
| C32 | 73 | M | Cecum | 1X0.5X0.5 | Adenocarcinoma |
| C33 | 57 | M | Sigmoid colon | 0.7X0.6X0.1 | Adenocarcinoma |
| C34 | 58 | F | Sigmoid colon | 0.6X0.5X0.2 | Adenocarcinoma |
| C35 | 71 | F | Rectum | 1.5X1X0.2 | Adenoma |
| C36 | 62 | M | Transverse colon | 0.8X0.5X0.2 | Adenocarcinoma |
| C37 | 78 | M | Rectum | 0.3X0.2X0.1 | Adenocarcinoma |
| C38 | 78 | F | Sigmoid colon | 0.3X0.2X0.1 | Adenocarcinoma |
| C39 | 53 | F | Sigmoid colon | 0.3X0.2X0.1 | Adenocarcinoma |
| C40 | 66 | F | Rectum | 0.3X0.2X0.1 | Adenocarcinoma |
Abbreviations: F, female; M, male.
4.2. Expression Level of Target Genes
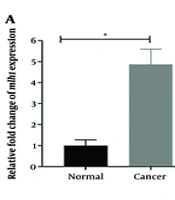
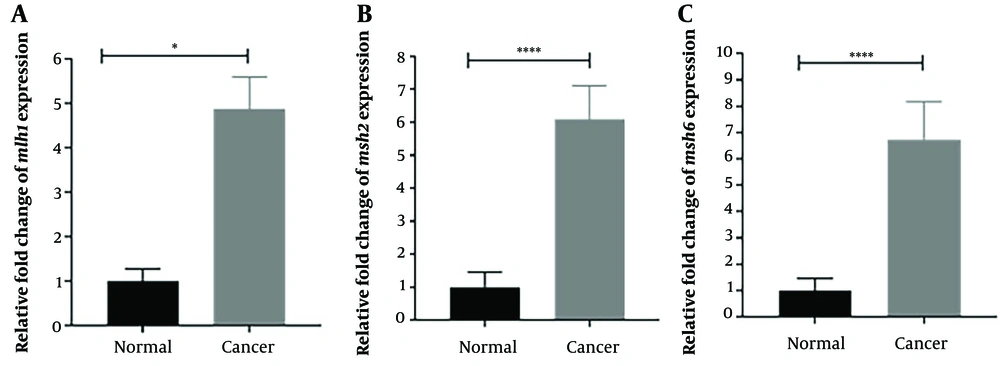
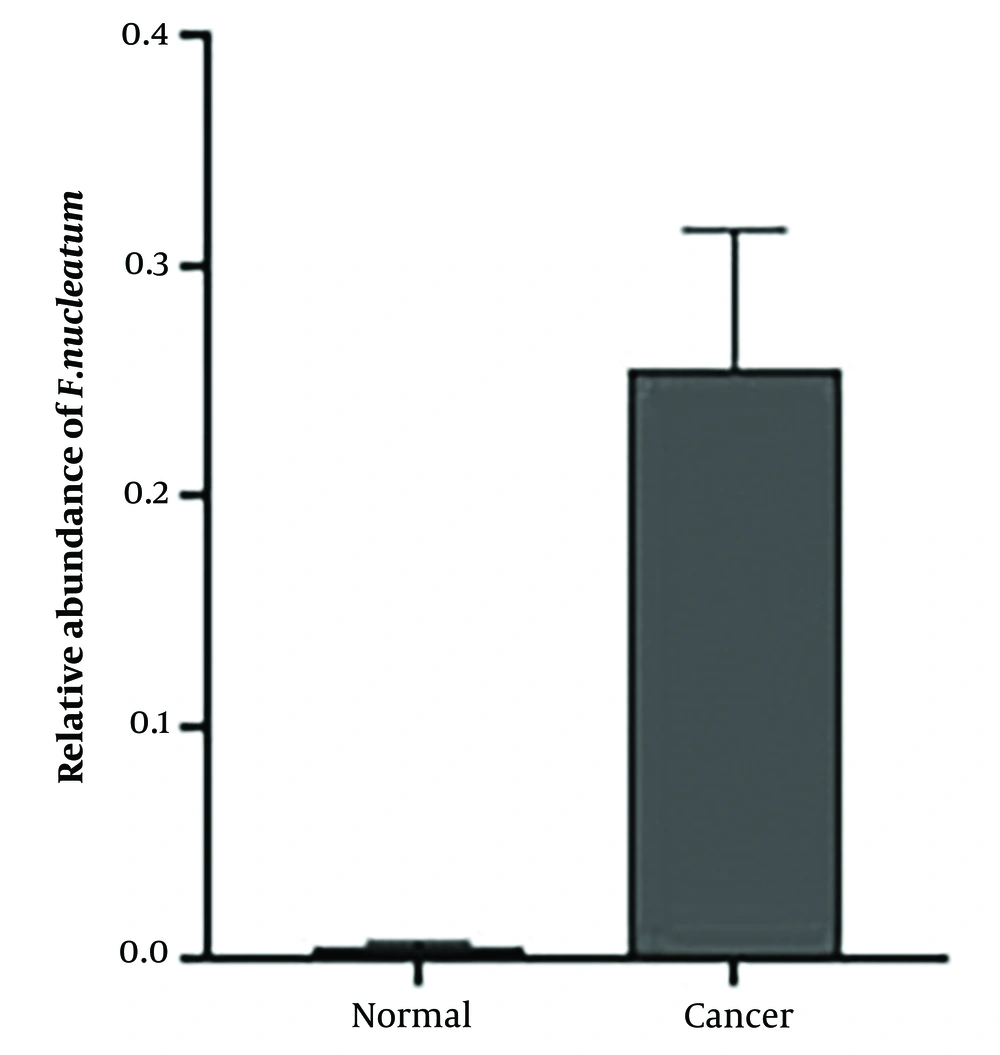
The relative expression levels of the mlh1, msh2, and msh6 genes, assessed using real-time PCR with gapdh as the internal control, showed a significant increase (P < 0.05) in the cancer group compared to the control group. The fold change analysis revealed a 5-fold increase in mlh1 expression, a 6.5-fold increase in msh2 expression, and a 7-fold increase in msh6 expression in the cancer group compared to the control group (Figure 1). To determine the relative abundance of F. nucleatum, real-time PCR with 16srRNA gene primers specific to F. nucleatum was employed. The results indicated a significantly higher frequency of F. nucleatum in the cancer group compared to the control group (P < 0.05) (Figure 2).
4.3. Abundance of Fusobacterium nucleatum in Cancer and Control Groups
The relative frequency of F. nucleatum was significantly higher in cancer patients (70%) compared to healthy individuals (25%). Additionally, F. nucleatum was more prevalent among female cancer patients (58%) than male patients (42%). Females aged 70 - 80 and males aged 50 - 80 showed a higher presence of F. nucleatum. A correlation was found between the tumor location and the frequency of F. nucleatum; it was detected more frequently in tumors located in the distal part of the colon (63%) than in the proximal colon and rectum (27%). The prevalence of F. nucleatum was also higher in adenocarcinomas (71%) compared to adenomas (13%). The abundance of F. nucleatum varied across different sections of the large intestine, with the highest levels found in the sigmoid, followed by the rectum, cecum, ascending colon, hepatic flexure, descending colon, and transverse colon, respectively.
4.4. Relative Expression Levels of Selected Genes in the Presence and Absence of Fusobacterium nucleatum
In cancer groups, the expression levels of mlh1, msh2, and msh6 were higher in the presence of F. nucleatum than in its absence. Fold change analysis showed a 7.5-fold increase in the expression levels of msh2 and msh6 and an 8-fold increase in mlh1 expression in the presence of F. nucleatum.
5. Discussion
Global incidence estimates for CRC in 2020 were 19.8 per 100 000 people, with a higher rate in men (23.4) compared to women (16.2) (1). In Vietnam, from 1996 to 2015, 12 938 individuals were diagnosed with CRC, 53.9% of whom were men, and the mean age at diagnosis was consistently 60.0 years (14). In Iran, Zare-Bandamiri et al. reported that 57.4% of CRC patients were men, with a mean age of 55.8, and 45.5% of patients fell within the 50 - 70 age range (15). Another study in Iran showed that of 562 CRC patients, 39% had early-onset CRC (under 50 years old) and 60% had late-onset CRC (over 50 years old), with participant ages ranging from 20 to 90 years and an average age of 55.63 years (16). The higher occurrence of CRC in men, as observed in our study, could be influenced by factors such as sample size, study duration, and regional differences. Interestingly, CRC was less common in the proximal area of the colon, aligning with findings from several Middle Eastern studies (17-19).
Differences in gene expression and tumor phenotype between proximal and distal lesions may explain the varying mechanisms of tumor progression. Proximal lesions, often smaller in size, might be more frequently overlooked during colonoscopy than distal lesions (20, 21). Our study did not reveal any significant differences in age or tumor location across age groups, suggesting a similar distribution of distal tumors in both CRC groups (22-24). However, it's important to note that other studies have reported conflicting results on tumor localization, continuing the debate on this issue (25, 26). Similar findings were reported by Ramsoekh et al., who noted an older age at CRC onset among male carriers of msh6 and mlh1 mutations, as well as considerable variation in the age of CRC onset between carriers of msh6 and msh2 mutations (48 vs. 43 years) (27).
Ulreich et al. studied 165 individuals with CRC, finding that 86.6% had mlh1-proficient CRCs, and 13.3% had mlh1-deficient CRCs (28). Engel et al. reported that individuals with pathogenic msh2 mutations had a 10% chance of developing advanced adenoma, compared to a 7.7% risk among those with mlh1 mutations. Moreover, a higher percentage of patients with pathogenic mutations in mlh1 or msh2 developed CRC within 10 years (11.3% and 11.4%, respectively) compared to those with msh6 mutations (29). However, other studies have associated the mlh1/msh2 phenotype with CRC (30, 31), suggesting that genetic variations may indirectly increase the risk of MSI-H CRC.
Our study found that 70% of cancer patient samples and 25% of control group samples were infected with F. nucleatum. The bacterium was predominantly found in the distal part of the colon, especially in the sigmoid and rectum, which are commonly associated with adenocarcinoma morphology. This aligns with a study that found a higher association of tumors in the distal part of the colon with F. nucleatum (32), suggesting that tumor development in these locations could be related to F. nucleatum colonization. However, other studies have reported varying results, with some indicating a preference for F. nucleatum colonization in the proximal colon over the distal colon (33, 34). Furthermore, while some studies report higher F. nucleatum distribution in the rectum compared to the distal sigmoid, others have indicated the opposite (35-37). A systematic review and meta-analysis by Idrissi Janati et al. supported F. nucleatum infection in the colon as a risk factor for CRC (38). Additionally, Tahara et al. discovered F. nucleatum in 74% of tumor tissues from 149 CRC patients (39).
The results of this study align with a review and meta-analysis that identified a strong correlation between increased F. nucleatum expression in CRCs and mlh1 hypermethylation (40). Furthermore, research data has linked the quantity of F. nucleatum DNA in fresh-frozen CRC tissue with proximal tumor sites, greater depth of invasion, poorly differentiated tumors, and decreased expression of mismatch-repair proteins mlh1, msh2, and pms2 (40). Studies also revealed that CRCs adjacent to normal colorectal tissues enriched with Fusobacterium were 15 times more likely to be Fusobacterium-enriched than CRCs close to normal Fusobacterium-free colorectal tissues (41).
The primary cause of MSI-H is deficits in MMR genes, including msh2, mlh1, and msh6 (42). According to our results, several studies have shown that F. nucleatum promotes CRC carcinogenesis in animal models, stimulating CRC cell development through E-cadherin/β-catenin signaling via the FadA adhesin, among other virulence components linked to CRC (43, 44). Immune evasion and/or chemoresistance due to F. nucleatum may explain the poor prognosis of F. nucleatum-associated CRC, potentially involving a complex relationship between CIMP/MSI and F. nucleatum infection mediated by ROS and nucleotide excision repair processes (45).
5.1. Conclusions
In total, our study provides compelling evidence supporting the association between F. nucleatum and CRC development and its potential role in poor prognosis and chemoresistance. The findings highlight the importance of F. nucleatum as a potential molecular marker for predicting CRC development. The dysregulation of critical genes involved in CRC pathogenesis due to F. nucleatum infection further supports the bacterium's direct impact on cancer development. These findings contribute valuable insights into the role of F. nucleatum in CRC and pave the way for potential targeted therapies and predictive markers for this disease. However, additional research is necessary to fully elucidate the underlying mechanisms and validate these findings in larger and more diverse cohorts.
There are several limitations to our study that need to be acknowledged and taken into account. Firstly, the sample size, particularly within the CRC group, was relatively small, potentially affecting the generalizability of our findings. Secondly, the absence of access to CRC grades posed a significant challenge, impairing our ability to assess the relationship between gene expression and cancer grade. Thirdly, our study could have benefited from conducting additional molecular evaluations and gene expression analyses to delve deeper into the mechanisms underlying F. nucleatum contribution to gastrointestinal damage. Addressing these limitations in future research endeavors would undoubtedly enhance the comprehensiveness and robustness of the investigations.