1. Background
Staphylococcus aureus is a major bacterial pathogen known for causing both uncomplicated skin infections and severe invasive infections worldwide. The mortality rate from S. aureus bacteremia is believed to exceed those of tuberculosis, human immunodeficiency virus, and hepatitis B. Additionally, methicillin-resistant Staphylococcus aureus (MRSA) presents a significant threat, causing severe infections in healthcare-associated (HAI) and community-acquired (CAI) settings. Methicillin-resistant S. aureus is characterized by the presence of the mecA gene, which codes for the PBP2a protein located within a specific region of the chromosome known as the Staphylococcal cassette chromosome mec (SCCmec) (1-3).
Panton-Valentine leukocidin (PVL) is a toxin produced by some S. aureus strains. The presence of PVL in these strains can influence the severity and outcome of an infection. Infections caused by PVL-associated S. aureus (PVL-SA) are commonly linked with community-acquired methicillin-resistant S. aureus (CA-MRSA) strains. However, the potential for nosocomial transmission poses a significant public health risk, as the presence of a PVL-positive clonal lineage of hospital-acquired MRSA (HA-MRSA) can rapidly spread, leading to more severe outcomes for infected patients (4).
The accessory gene regulator (agr) system is a global regulatory system in S. aureus that controls the expression of numerous virulence factors and regulatory molecules. This system comprises four genes (agrA, agrB, agrC, and agrD) that encode a quorum-sensing system responsible for coordinating the expression of virulence factors in response to cell density. The agr system plays a crucial role in regulating various virulence factors, including toxins, enzymes, and surface proteins, which contribute to the bacterium's pathogenicity. Recent studies have revealed a more complex understanding of the accessory gene regulator (agr) in S. aureus infection (5).
Molecular typing methods in bacteria refer to a set of techniques used to characterize and differentiate bacterial strains based on their genetic makeup. These methods enable researchers to identify genetic variations within bacterial populations, track the spread of specific strains, and understand the epidemiology of bacterial infections. Common molecular typing methods include pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), whole-genome sequencing (WGS), and polymerase chain reaction (PCR)-based techniques targeting specific genes or genetic elements (6). MLST for S. aureus involves sequencing several housekeeping genes to determine the sequence type (ST) of the bacterium. This information is crucial for tracking the spread of specific strains and understanding the epidemiology of S. aureus infections (7).
2. Objectives
In this research project, our objective was to analyze the antibiotic resistance profiles and genetic relatedness of community-associated (CA) and hospital-associated (HA) MRSA, along with methicillin-sensitive S. aureus (MSSA) strains obtained from patients in both hospital and community environments in Bandar Abbas, located in southern Iran. To achieve this, we employed SCCmec typing and MLST techniques for strain characterization. Additionally, we aimed to explore the role of the accessory gene regulator (agr) genes in these clinical S. aureus isolates.
3. Methods
This study was conducted as a cross-sectional investigation from 2020 to 2021, during which a total of 156 strains of S. aureus were collected from both community (outpatients) and hospital (inpatients) settings in Bandar Abbas, southern Iran. The S. aureus strains were sourced from patients who had been hospitalized for a minimum of 72 hours, excluding duplicate samples from the analysis. Confirmation of S. aureus isolates was performed through gram staining and biochemical tests, including growth evaluation on blood agar medium, hemolysis, catalase activity, mannitol fermentation, and coagulase and DNase tests (8). Once identified, the S. aureus isolates were preserved for subsequent procedures using Trypticase Soy Broth (TSB) containing 30% glycerol.
3.1. Antimicrobial Susceptibility Testing
The susceptibility of S. aureus isolates to various antibiotics, such as azithromycin (15 μg), tigecycline (15 μg), gentamicin (10 μg), linezolid (30 μg), clindamycin (2 μg), ciprofloxacin (5 μg), and tetracycline (30 μg), was assessed using the Kirby-Bauer disc diffusion method following the Clinical and Laboratory Standards Institute (CLSI 2021) guidelines (9). The reference strain S. aureus ATCC25923 served as the control. Methicillin resistance was determined using a cefoxitin disc (FOX, 30 μg) according to the CLSI guidelines. The multidrug-resistant (MDR) phenotype was defined as acquired non-susceptibility to at least one agent in three or more antibiotic categories, and pandrug-resistance (PDR) was defined as non-susceptibility to all agents in all antibiotic classes (10).
3.2. Extraction of DNA and Amplification of Genes
The genomic DNAs from all S. aureus isolates were extracted using a previously established protocol. This process involved dissolving 20 μL of buffer containing 0.25% SDS (sodium dodecyl sulfate) and 0.05 M NaOH in 200 mL of deionized water, which was then placed into a microtube. Fresh bacterial colonies were dissolved in this solution and heated at 95°C for 10 minutes. Subsequently, the solution was centrifuged at 13,000 g for 1 minute, and 180 μL of deionized water was added to it (11). To identify MRSA strains, all S. aureus isolates were screened for the presence of the mecA gene through PCR assays conducted using a SensoQuest Labcycler thermal cycler, following the method outlined by Mahmoudi et al. (12). Additionally, the agr and pvl genes were also detected using PCR with specific primers (13). PCR reactions were prepared with a total volume of 25 μL. The presence of various SCCmec types (I-V) among S. aureus isolates was investigated using specific primers designed for SCCmec types and subtypes I, II, III, IV, and V (14).
3.3. Multilocus Sequence Typing
Five S. aureus isolates were selected for further characterization using MLST. The MLST for S. aureus was conducted following the scheme proposed by Enright et al (15). Internal fragments of seven housekeeping genes (arcC, gmk, aroE, glpF, pta, tpi, and yqil) were amplified using specific primers and conditions outlined in the online MLST database (https://pubmlst.org) and then subjected to direct sequencing. Allele numbers and sequence types (STs) were assigned based on the guidelines provided in the S. aureus typing section of the PubMLST database.
3.4. Statistical Analysis
The statistical software used for data analysis was SPSS version 22. Fisher's exact test was employed to determine significant associations between categorical variables. A significance level of P < 0.05 was considered significant.
4. Results
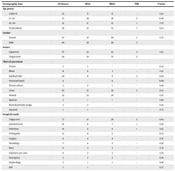
In this cross-sectional study, a total of 156 S. aureus-positive culture samples were collected from both outpatients and inpatients. Of these, 87 (56%) isolates were from female patients and 69 (44%) were from male patients, with ages ranging from under 15 to over 65 years. Demographic information for MRSA and MSSA is presented in Table 1. A total of 156 S. aureus strains were isolated, including 79 from inpatients (hospital-acquired: HA) and 77 from outpatients (community-acquired: CA). Among these, 70 (44.8%) were identified as MRSA based on both disc diffusion and PCR results. Of the MRSA isolates, 45 of 97 (46.3%) were from inpatients and 25 of 59 (42.3%) were from outpatients. Table 1 also compares MRSA and MSSA strains based on the sources of clinical samples and places of sampling (hospital wards). The statistical analysis showed no significant differences in age, gender, type of clinical sample, or hospitalization versus outpatient status between MSSA and MRSA strains. However, a significant difference was observed between MSSA and MRSA strains only in the orthopedic unit and other hospital units.
| Demographic Data | All Strains | MSSA | MRSA | PDR | P-Value |
|---|---|---|---|---|---|
| Age group | |||||
| Under 15 | 22 | 9 | 13 | - | 0.14 |
| 15 - 44 | 74 | 46 | 28 | 2 | 0.09 |
| 45 - 64 | 32 | 17 | 15 | 1 | 0.79 |
| 65 and above | 28 | 14 | 14 | 1 | 0.54 |
| Gender | |||||
| Female | 87 | 47 | 40 | 2 | 0.75 |
| Male | 69 | 39 | 30 | 2 | |
| Source | |||||
| Inpatients | 97 | 52 | 45 | 2 | 0.62 |
| Outpatients | 59 | 34 | 25 | 2 | |
| Clinical specimens | |||||
| Ascites | 2 | 2 | - | - | 0.42 |
| Blood | 11 | 8 | 3 | - | 0.22 |
| Tracheal tube | 20 | 11 | 9 | 2 | 0.99 |
| Personnel hand | 8 | - | 8 | - | 0.00 |
| Throat culture | 3 | 2 | 1 | - | 0.68 |
| Urine | 65 | 37 | 28 | 2 | 0.70 |
| Wound | 42 | 22 | 20 | - | 0.67 |
| Sputum | 2 | 1 | 1 | - | 0.88 |
| Bronchoalveolar lavage | 2 | 2 | - | - | 0.42 |
| Synovial | 1 | 1 | - | - | 0.77 |
| Hospital wards | |||||
| Outpatient | 77 | 41 | 36 | 2 | 0.64 |
| Internal ward | 18 | 11 | 7 | 1 | 0.58 |
| Infectious | 14 | 5 | 9 | 1 | 0.12 |
| Orthopedic | 13 | 11 | 2 | - | 0.02 |
| Surgery | 8 | 4 | 4 | - | 0.76 |
| Neurology | 7 | 4 | 3 | - | 0.91 |
| Burn | 6 | 3 | 3 | - | 0.79 |
| Intensive care unit | 6 | 3 | 3 | - | 0.79 |
| Emergency | 3 | 2 | 3 | - | 0.68 |
| Nephrology | 2 | 1 | 1 | - | 0.88 |
| ENT | 1 | 1 | - | - | 0.77 |
Abbreviations: AZT, azithromycin; CIP, ciprofloxacin; CLI, clindamycin; GEN, gentamicin; LZD, linezolid; MRSA, methicillin-resistant S. aureus; MSSA, methicillin-sensitive S. aureus; TE, tetracycline; TIG, tigecycline; PDR, pandrug-resistance.
Table 2 presents the results of antimicrobial susceptibility testing against seven different antibiotics. According to disc diffusion results, the highest resistance was observed against azithromycin (83%) and the lowest was against linezolid (5%) and gentamicin (7%). Resistance rates were 44% for cefoxitin, 43% for tetracycline, 24% for clindamycin, 10% for ciprofloxacin, and 9% for tigecycline. All MRSA strains exhibited the MDR phenotype, and 4 out of 70 (5.7%) MRSA strains showed the pandrug-resistant (PDR) phenotype.
Table 3 also compares antibiotic resistance patterns between MRSA and MSSA strains, revealing significant differences in resistance to tetracycline, linezolid, and azithromycin between the two strains. Table 3 additionally displays the prevalence of various types of pvl and SCCmec genes in S. aureus isolates from hospital and community settings. The PCR results revealed that the pvl gene was present in three isolates: Two hospital-acquired (with SCCmec type IV and III and agr type III, I) and one community-acquired (with agr type I). The hospital strains were isolated from synovial fluid and bronchoalveolar lavage fluid samples, while the CA strain was isolated from a urine sample. The mec gene was identified in the PVL-positive strain obtained from bronchoalveolar lavage fluid, classifying it as an MRSA strain.
| Antibiotics | |||||||
|---|---|---|---|---|---|---|---|
| AZT | TIG | GEN | LZD | CLI | CIP | TE | |
| Inpatient (n = 79) | MSSA: 19 (4) | MSSA: 4 (5) | MSSA: 4 (5) | MSSA: 6 (7.5) | MSSA: 8 (10) | MSSA: 5 (8.4) | MSSA: 13 (16.4) |
| MRSA: 17 (21) | MRSA: 2 (2.5) | MRSA: 2 (2.5) | MRSA: 1 (1.3) | MRSA: 9 (11.3) | MRSA: 4 (5) | MSSA: 13 (16.4) | |
| Outpatient (n = 77) | MSSA: 26 (34) | MSSA: 2 (2.5) | MSSA: 2 (2.5) | MSSA: 1 (1.2) | MSSA: 10 (13) | MSSA: 4 (5) | MSSA: 13 (16.8) |
| MRSA: 21 (27) | MRSA: 5 (6.4) | MRSA: 4 (5) | MSSA: 0 (0) | MRSA: 10 (13) | MRSA: 2 (2.5) | MRSA: 28 (36) | |
| Total | 83 (53) | 13 (9) | 12 (7) | 8 (5) | 37 (24) | 15 (10) | 67 (43) |
| P-value | 0.01 | 0.36 | 0.44 | 0.02 | 0.38 | 0.15 | 0.01 |
Abbreviations: TE, tetracycline; TIG, tigecycline; AZT, Azithromycin; CIP, ciprofloxacin; CLI, clindamycin; GEN, gentamicin; LZD, linezolid; MRSA, methicillin-resistant S. aureus; MSSA, methicillin-sensitive S. aureus; PDR, pandrug-resistance.
a Values are expressed as No. (%).
| Inpatient (n = 79) | Outpatient (n = 77) | Total (n = 156) | P-Value | |
|---|---|---|---|---|
| mecA gene | 34 (43) | 36 (47) | 70 (45) | 0.32 |
| pvl | 2 (2.5) | 1 (1) | 3 (2) | 0.3 |
| Agr I | 19 (24) | 6 (8) | 25 (16) | 0.005 |
| Agr II | 4 (5) | 10 (13) | 14 (9) | 0.07 |
| Agr III | 9 (11) | 1 (1) | 10 (6) | 0.005 |
| Agr IV | 1 (1) | 0 (0) | 1 (0.6) | 0.34 |
| Agr I, III, IV | 1 (1) | 0 (0) | 1 (0.6) | 0.34 |
| Sccmec I | 5 (6) | 4 (5) | 9 (6) | 0.38 |
| Sccmec II | 3 (4) | 0 (0) | 3 (2) | 0.09 |
| Sccmec III | 10 (13) | 6 (8) | 16 (10 ) | 0.15 |
| Sccmec IV | 6 (8) | 1 (1) | 7 (4.5) | 0.02 |
| Sccmec IV, III | 3 (4) | 0 (0) | 3 (2) | 0.09 |
a Values are expressed as No. (%).
Table 3 presents the frequency of virulence genes and molecular typing of MRSA isolates. The PCR results for agr genes among strains from both hospital and community settings indicated that S. aureus isolates from inpatients and outpatients encompassed four distinct agr types. Among these, agr type I had the highest number of isolates originating from hospital settings. The findings also revealed that agr types I and III were more prevalent in strains isolated from hospitals compared to those from the community. However, the occurrence of the agr II group was higher in community isolates than in hospital isolates.
A significant difference was observed in the frequency of agr group I and agr group III between hospital-associated (HA) and community-associated (CA) isolates. Only one hospital strain exhibited the simultaneous presence of agr types I, III, and IV. The results of SCCmec typing indicated that SCCmec III was the most prevalent among the SCCmec types. The frequency of different SCCmec types was higher in hospital isolates than in community isolates. Statistical analysis revealed a significant difference in the frequency of SCCmec IV between hospital and community isolates.
The MLST results revealed that the five MRSA strains examined have new STs: ST8634, ST8640, ST8650, ST8651, and ST8652, which are being introduced and reported for the first time in this study. The number of alleles and the assigned STs for these five strains are listed in Table 4. The MLST findings demonstrate genetic diversity in MRSA strains isolated from both hospital and community settings. None of these identified STs belonged to clonal complexes; instead, they were considered singletons or unique STs.
| Strain | In/Out | Source | Sequence Type | arcC Allele | aroE Allele | glpF Allele | gmk Allele | Pta Allele | tpi Allele | ygiL Allele |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Out | Urine | 8634 | 853 | 37 | 19 | 89 | 805 | 665 | 32 |
| 6 | In | Wound | 8640 | 12 | 4 | 1 | 437 | 1 | 724 | 87 |
| 15 | In | Trachea | 8650 | 3 | 1 | 319 | 475 | 787 | 8 | 879 |
| 24 | In | Blood | 8651 | 620 | 653 | 1 | 78 | 504 | 698 | 96 |
| 44 | In | Trachea | 8652 | 79 | 1 | 14 | 23 | 691 | 497 | 398 |
5. Discussion
This study evaluated antibiotic resistance, the prevalence of genes such as mecA, pvl, agr, SCCmec, and the genetic relatedness between MRSA and MSSA strains in isolates from healthcare-associated and community-associated infections. Our findings indicate that 43.6% of both outpatients and inpatients in Bandar Abbas, southern Iran, were infected with MRSA. This rate aligns with other studies conducted in Iran, underscoring the significance of MRSA presence in the community, which exceeds 40% and demands special attention for control and management from a public health perspective.
Analysis of multiple studies in Iran shows that the rate of MRSA infections among confirmed S. aureus isolates is approximately 43.0%. Detailed analyses have shown that the prevalence of MRSA was higher in studies conducted after 2000 (16). The high prevalence of MRSA in healthcare settings in Iran could be attributed to various factors such as indiscriminate antibiotic use, insufficient implementation of prophylactic hygiene measures, inadequate staff training, and lack of hospital infection control programs. Furthermore, the association of multidrug resistance with MRSA has compounded the challenges of managing MRSA in hospitals across Iran. issue the prevalence of MRSA in Iran, at 43.0%, does indeed appear to be higher than rates reported in other studies (16).
The overall prevalence of clinically isolated MRSA in Egypt is notably high at 63% (17). Variations in MRSA prevalence across different regions of Iran may be due to differences in infection control practices, healthcare conditions, antibiotic prescription practices, selective antibiotic pressure in hospitals, and underlying clinical conditions. These factors can contribute to the varying rates of MRSA prevalence in different regions of the country (18).
Recent evidence indicates that CA-MRSA accounts for a significant portion of all MRSA infections, with these strains also being detected in hospitalized patients, suggesting intra-hospital movement of CA-MRSA. The prevalence of CA-MRSA strains varies by country, with the lowest reported in France and the highest in the United States (19). In our study, the frequency of CA-MRSA strains was 42%, aligning with results reported from Tehran, Shiraz, and Hamadan in Iran (20-22). However, some studies in Iran have reported lower frequencies of MRSA than those found in our study (18, 23). No significant differences were observed between the prevalence of CA-MRSA and HA-MRSA strains in our study. CA-MRSA outbreaks and cases have been reported in various countries in the Asia-Pacific region, indicating that these strains are circulating in the region (24). High levels of resistance to azithromycin, cefoxitin, and tetracycline were found in both HA-MRSA and CA-MRSA isolates.
Based on the results of this study, linezolid, gentamicin, and tigecycline are the most effective antibiotics against MSSA and MRSA strains acquired from both community and hospital settings. Linezolid is effective against more than 98% of Staphylococcus infections, with resistance detected in only 0.05% of S. aureus infections. In many studies conducted in Iran, resistance to linezolid was either not reported or was very low (25). Studies analyzing the resistance of Staphylococcus isolates to linezolid in various countries found that the United States, Canada, and European countries had higher levels of resistance, while African and Asian countries reported the lowest (0.1%) resistance among MRSA strains (26).
The 5% linezolid resistance observed in our study is significant, although resistance was higher in hospital strains than in community strains. Consistent with our findings, studies from Egypt and Iran reported 5% linezolid resistance in MRSA isolates (26). A recent study conducted in Pakistan found that 35% of MRSA strains were resistant to linezolid (27). The presence of linezolid resistance in hospital isolates is higher than in the community, as this antibiotic is primarily prescribed in hospitals, but the emergence of resistance in community isolates is notable and concerning.
Tigecycline demonstrated favorable efficacy, following linezolid and gentamicin, against MRSA and MSSA strains in both hospital and community settings. The geographical variation in resistance to tigecycline compared to linezolid suggests different levels of antibiotic usage across regions. Despite recommendations for treating skin and soft tissue infections, recent MRSA infection treatment guidelines have not yet included tigecycline. Previous studies suggest that there are no significant differences between tigecycline and other newer medications, positioning tigecycline as a secondary or tertiary treatment option for MRSA-related infections (28, 29). Our results align with previous studies conducted in Iran, where we found SCCmec type III to be the predominant type among both hospital-acquired and CA MRSA isolates (18, 30-32).
The prevalence of SCCmec type III and type 3 ccr among MRSA strains from various sources, including hospitals, the environment, and animals, suggests the hospital origin of MRSA isolates in Iran (18). Additionally, our study detected other SCCmec types such as I, IV, and II, aligning with findings from other studies in Iran. A study from western Iran did not detect SCCmec V (33). However, some studies from Iran have reported the presence of S. aureus strains with SCCmec V (18, 30, 34). In our study, SCCmec type IV was found in both hospital-acquired (HA) and CA isolates. The detection of one isolate with SCCmec IV in a hospital suggests the potential dissemination of MRSA strains from the community to hospitals. A study from Japan indicated that the prevalence of SCCmec IV isolates, which are primarily CA MRSA, has increased in Japanese hospitals (35). The prevalence of pvl gene-carrying isolates varies, but it has been reported with a higher incidence in community-associated MRSA strains. In our study, the prevalence of pvl gene-carrying isolates was 1.9%, which is lower than in most Iranian studies, where the prevalence ranged from 22.7% to 52.9% (36).
CA-MRSA appears to be associated with increased transmission and hospitalization, as well as skin and soft tissue infections such as furuncles, cellulitis, and skin abscesses. Rarely, it can lead to severe diseases such as necrotizing pneumonia (36). In our study, the PVL-positive strain isolated from the community was obtained from a urine sample, and PVL-positive HA strains were isolated from synovial fluid and bronchoalveolar lavage fluid. Consistent with our findings, a study from Iran reported that among 26.3% of PVL-positive strains were HA-MRSA that presumably moved to the community (37). Additionally, a recent study from two hospitals in Greece conducted between 2020 and 2022 found that 19.15% of the isolates were PVL-positive (38). The presence of PVL-positive strains in hospital isolates indicates the transfer of CA-MRSA from the community to the hospital. A systematic surveillance program is needed to identify common PVL-positive clones in the community.
In this study, it was found that hospital isolates contained all four agr groups (agr I, agr II, agrIII, and agr IV), while CA strains lacked the agr IV group. Consistent with many other studies, agr group I was the predominant group in hospital-acquired infection (HAI) isolates. However, agr II was the predominant agr group in CA strains (39). Interestingly, no significant difference was observed in the frequency of agr groups between HA and CA strains. This suggests that the distribution of agr groups may not be a distinguishing factor between hospital-acquired and CA strains. The discovery of five new typing sequences in S. aureus isolates is an interesting finding. These new sequences were reported for the first time and recorded in the PubMLST database, indicating the genetic diversity and circulation of different bacterial clones in and outside hospitals. These unique STs did not belong to any clonal complex of S. aureus, underscoring the need for further studies to understand their origins and potential implications in both hospital and community settings.
The evidence indicating the predominance of the ST239 hospital-acquired MRSA clone in many Asian countries, including Iran, is noteworthy. In the study by Bourbour et al., a significant proportion of MRSA strains isolated from inpatients in a teaching hospital in Tehran belonged to ST239 (50%), with ST30 detected in 30% of isolates (40). In studies conducted in hospitals in Isfahan and Tehran, Iran, it was found that 47% and 72% of clinical MRSA strains belonged to ST239, respectively (41, 42). This highlights the significant prevalence of the ST239 hospital-acquired MRSA clone in these regions, emphasizing the importance of understanding and addressing this particular strain in healthcare settings.
This data underscores the importance of continuous monitoring and focused efforts to control and prevent the spread of MRSA in these regions. Further research is needed to understand the epidemiology of MRSA strains, including the newly identified STs in this study. This will aid in developing effective strategies to control and prevent the spread of these bacteria in healthcare settings and the community. The limitation of financial resources in the study prevented the extensive use of MLST to determine the STs for a larger number of strains. This constraint may have impacted the comprehensive understanding of the genetic diversity and distribution of bacterial clones in hospital and community settings.
5.1. Conclusions
In conclusion, the dissemination of HA-MRSA isolates to the community represents a significant public health concern. This underscores the importance of implementing effective processes to control the spread of isolates from hospitals to communities and vice versa. It is crucial to establish and maintain robust infection control measures, surveillance systems, and communication channels between healthcare facilities and community health organizations to prevent the transmission of MRSA and other antibiotic-resistant bacteria. A proactive approach is essential for protecting public health and reducing the impact of antibiotic-resistant infections in both healthcare and community settings. Identifying the sources of infection in both hospital and community settings is vital for the effective prevention and control of antibiotic-resistant bacteria such as MRSA. Understanding the origins and pathways of transmission can guide the development of targeted interventions and control measures.