1. Background
Catheter-associated urinary tract infections (CAUTIs) are common in healthcare-associated infections (1). Around 40% of healthcare-associated infections are urinary tract infections, with about 80% of cases attributed to the use of urinary catheters (2). The use of urinary catheters ranges from 12% to 16% among adult hospital patients, and the incidence of CAUTIs ranges from 3% to 7% in these patients (3). The total number of urinary tract infections worldwide is 222 million (3). Diagnosing CAUTIs relies on both clinical and laboratory diagnoses of bacteriuria. Signs and symptoms of these infections include fever, suprapubic tenderness, and flank pain (4, 5). Asymptomatic bacteriuria alone, without predisposing risk factors for infections, does not indicate antibiotic therapy (6). The removal of the catheter, along with antibiotics and other drugs, must be indicated by the clinician (7, 8). Infection control guidelines for preventing CAUTIs depend on knowledge of the data concerning risk factor analysis of CAUTIs (9).
The most common pathogen associated with urinary tract infections is Escherichiacoli, responsible for about 90% of community-acquired infections and around 50% of healthcare-associated infections (3). Various virulence factors in uropathogenic E. coli facilitate the invasion, growth, and persistence in the urinary tract epithelium (10). The host defense mechanisms include cytokine secretion such as interleukin 8, uroepithelial defensin, cytopeptide expression like Tamm-Horsfall protein, and urinary flow (11). The formation of biofilm by E. coli represents a virulence mechanism where microbial cells are irreversibly bound to the epithelial surface with the formation of polysaccharide material (12). The biofilm prevents the access of antimicrobial agents and antibodies to the bacterial cells and provides nutrients to the bacterial cells (13).
Various virulence genes in uropathogenic E. coli increase the severity of urinary tract infections. These genes include P fimbriae (pap), type 1-fimbriae (fim-H), fimbrial-adhesin1 (afa1), S-fimbriae (sfa), hemolysin (hly), and cytotoxic necrotizing factor (cnf1) (14). These genes are responsible for colonizing the host epithelium, evading host mechanisms, and invading with injuries to host cells (15). There is a link between the presence of these virulence genes, biofilm formation, and antibiotic resistance in E. coli. These factors need careful assessment to avoid antibiotic therapy failure (16). Despite the known role of these virulence factors in the severity of urinary tract infections, few studies have explored their association with CAUTIs, particularly in the context of biofilm formation and antibiotic resistance. This represents a significant gap in the literature, as understanding these associations could inform more effective treatment strategies and help prevent antibiotic therapy failure.
2. Objectives
This study aims to address these gaps by evaluating the various risk factors associated with CAUTIs, with a particular focus on the capacity of E. coli to form biofilms and the presence of various virulence genes in these infections.
3. Methods
This retrospective study included patients with CAUTIs admitted to the intensive care units (ICU) of Mansoura University Hospital, Egypt, from January 2019 to March 2022. The inclusion criteria were patients aged >18 years diagnosed with healthcare-associated CAUTIs and inserted urinary catheters, according to the criteria identified by the centers for disease control (CDC) (17), with bacterial infection due to E. coli. The exclusion criteria were patients with community-acquired urinary tract infections and those with urinary tract infections not associated with catheters. The sample size depended on the non-probability sampling method and the time frame of the study, during which patients with CAUTIs due to E. coli were diagnosed.
3.1. Urine Sample
Urine samples were obtained from the urinary catheter and rapidly transported to the laboratory. A microscopic examination was carried out for pus cells, followed by a microbiological culture (18). Escherichia coli identification was performed using Gram stain and complete biochemical identification with triple sugar iron agar, Simmon citrate, urease, lysine, phenylalanine, Methyl red, Voges Proskauer test, and subculture on MacConkey agar (19). A pure suspension of E. coli was prepared by subculturing the isolates in brain heart infusion broth overlaid with glycerol, and the suspension was kept frozen at -20ºC for polymerase chain reaction (PCR) (20).
3.2. Antibiotics Sensitivity Test
An antibiotic sensitivity test was performed using the disc diffusion method. The antibiotics discs used were imipenem (10 μg), meropenem (10 μg), cefepime (30 μg), ceftriaxone (30 μg), gentamicin (10 μg), aztreonam (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), ciprofloxacin (5 μg), trimethoprim/sulfamethoxazole (25 μg), and nitrofurantoin (300 μg) (Oxoid, United Kingdom). Pure colonies of E. coli were suspended in Muller-Hinton broth, and half McFarland concentrations were prepared. The suspension was then spread on Muller-Hinton agar. Antibiotic discs were placed on the agar, and the plates were incubated at 37°C for 24 hours. The diameter of the inhibition zones around the antibiotic discs was measured, and the results were interpreted as sensitive or resistant according to the guidelines of the Clinical Laboratory Standards Institute (CLSI) (21).
3.3. Biofilm Formation
Escherichia coli were grown in tryptic soy broth for 24 hours at 37°C. Then, a 1:20 dilution of the cultures was performed in tryptic soy broth, and 200 microliters were added to each well in a 96-well microplate, with incubation for 48 hours at 37°C. The wells were stained with 2% crystal violet and incubated for 15 minutes. The wells were then washed with sterile distilled water, and 150 microliters of 95% ethanol was added to the wells. The microplate was read using a microplate reader at a wavelength of 570 nm (22).
3.4. Extraction of DNA
The DNA of E. coli isolates was extracted using the boiling method as previously described (23).
3.5. Amplification and Detection
The primers used are summarized in Table 1 and (24). The amplification mixture was prepared using a Qiagen ready-to-use mixture. The amplification procedure included the following steps using an Eppendorf Master Cycler: Initial denaturation at 94°C for 2 minutes, followed by thirty cycles of denaturation at 94°C for 60 seconds, annealing at 63°C for 30 seconds, and extension at 72°C for 90 seconds, with a final extension step at 72°C for 5 minutes. Sterile distilled water was used as a negative control. Detection was performed by electrophoresis on a 1.5% agarose gel stained with ethidium bromide for 20 minutes, followed by ultraviolet visualization of the bands.
| Gene | The Sequences of the Primers | Bp |
|---|---|---|
| PapC | F GACGGCACTGCTGCAGGGTGTGGCG | 328 |
| R ATATCCTTTCTGCAGGGATGCAATA | ||
| Fim-H- | F TGTACTGCTGATGGGCTGGTC | 564 |
| R GGGTAGTCCGGCAGAGTAACG | ||
| Sfa- | F CTCCGGAGAACTGGGTGCATCTTAC | 410 |
| R CGGAGGAGTAATTACAAACCTGGCA | ||
| Afa | F GCTGGGCAGCAAACTGATAACTCTC | 750 |
| R CATCAAGCTGTTTGTTCGTCCGCCG | ||
| HlyA | F AACAAGGATAAGCACTGTTCTGGCT | 1177 |
| R ACCATATAAGCGGTCATTCCCGT | ||
| Cnf1 | F AAGATGGAGTTTCCTATGCAGGAG | 498 |
| R TGGAGTTTCCTATGCAGGAG |
3.6. Statistical Analysis
The data from the study was analyzed using SPSS 22. Qualitative data was expressed as numbers and percentages, with comparisons performed using the chi-square test. Significance was indicated by a P-value of < 0.05. Quantitative data was expressed as mean and standard deviation (SD).
4. Results
4.1. Demographic Characteristics and Clinical Data
The study included 204 patients with healthcare-associated CAUTIs. Among them, 107 (52.5%) were males and 97 (47.5%) were females, with a mean age of 49.53 ± 14.7 years. The common comorbidities were diabetes mellitus (DM) (33.3%), renal disorders (23.5%), and post-operative complications (11.3%). Various clinical conditions led to ICU admission, including common malignancy (24.0%), intracranial hemorrhage (20.6%), and renal failure (15.2%). Recurrent urinary tract infections were reported in 24 patients (11.4%). The most common clinical symptoms and signs were pain on clinical examination (55.4%), pain on micturition (46.6%), and fever (45.6%) (Table 2).
| Demographic and Clinical Data | Values |
|---|---|
| Sex | |
| Male | 107 (52.5) |
| Female | 97 (47.5) |
| Age (y) | (49.53 ± 14.79) |
| Comorbidities | |
| Diabetes mellitus | 68 (33.3) |
| Kidney diseases | 48 (23.5) |
| Post-operative complications | 23 (11.3) |
| Underlying etiology | 49 (24.0) |
| Malignancy | 19 (9.3) |
| Diabetic complications | 31 (15.2) |
| Renal failure intracranial hemorrhage | 42 (20.6) |
| Surgical complications | 22 (10.8) |
| Myocardial infarctions | 6 (2.9) |
| Hepatic disorders | 26 (12.7) |
| Miscellaneous | 9 (4.4) |
| Recurrent urinary tract infections | 24 (11.8) |
| Clinical signs and symptoms | |
| Fever | 93 (45.6) |
| Urgency | 48 (3.5) |
| Frequency | 22 (10.8) |
| Pain in the micturition | 95 (46.6) |
| Pain on examination | 113 (55.4) |
a Values are expressed as No. (%) or mean ± SD.
4.2. Biofilm Formation
The capacity to form biofilm in vitro was detected in 111 (54.4%) of the isolated E. coli strains.
4.3. Antibiotic Resistance
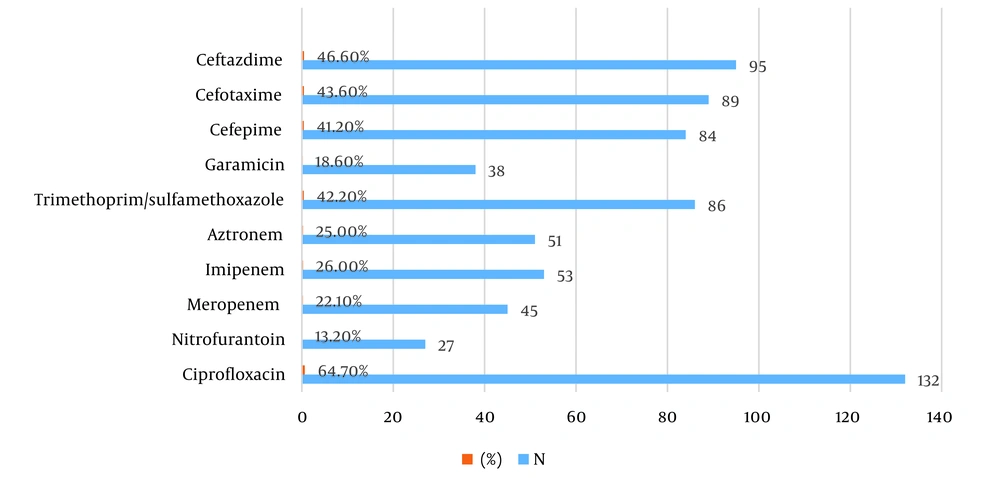
The most frequently detected resistances among isolated E. coli were to ciprofloxacin (64.7%), followed by resistance to the third-generation cephalosporin ceftazidime (46.6%) and cefotaxime (43.6%). Additionally, resistance to trimethoprim/sulfamethoxazole was observed in 42.2% of the isolates (Figure 1).
4.4. Virulence Gene Detection
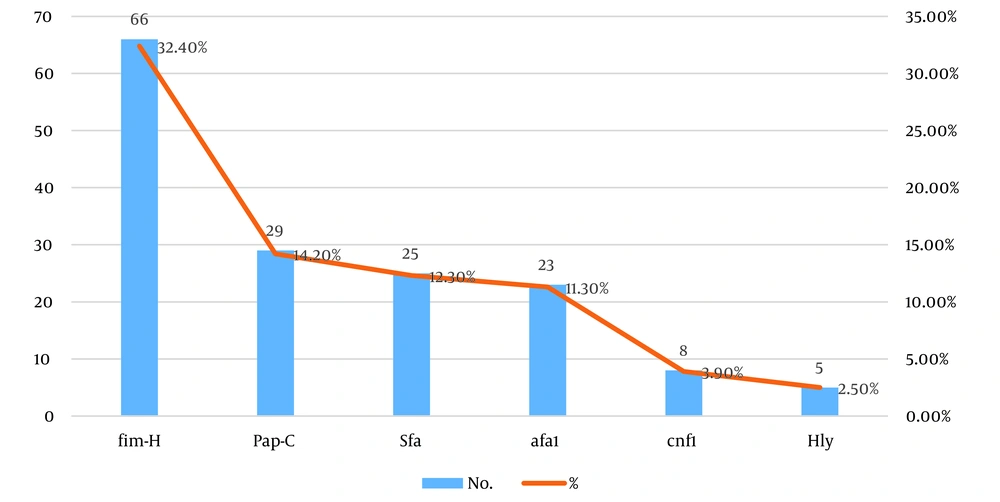
The most frequently detected genes were fim-H (32.4%), followed by pap-C (14.2%), sfa (12.3%), and afa1 (11.3%). The least frequently detected genes were cnf1 (3.9%) and hyl (2.5%) (Figure 2). There was a significant association between the presence of fim-H, pap-C, afa1, and hyl genes (P = 0.001, P = 0.001, P = 0.04, P = 0.046 respectively) and the capacity to form biofilm in isolated E. coli (Table 3).
| Virulence Genes | Positive Biofilm (n = 111) | Negative Biofilm (n = 93) | P-Value |
|---|---|---|---|
| fim-H | 64 (57.7) | 2 (2.2) | 0.001 |
| Pap | 25 (22.5) | 4 (4.3) | 0.001 |
| Sfa | 13 (11.7) | 12 (12.9) | 0.48 |
| Cnf | 2 (1.8) | 6 (6.4) | 0.1 |
| Afa-1 | 17 (15.3) | 6 (6.4) | 0.04 |
| Hyl | 5 (4.5) | 0 (0) | 0.046 |
| Cnf | 2 (1.8) | 6 (6.4) | 0.1 |
a Values are expressed as No. (%).
Urinary tract infections were most commonly detected 7 - 10 days after catheter insertion (48.53%), followed by 3 - 6 days after insertion (45.59%). The least frequent occurrence of urinary tract infections was more than 10 days after catheter insertion (5.86%). There was a significant association between the early presence of CAUTIs (from 3 to 6 days) and the presence of DM (P = 0.046) and postoperative complications (P = 0.046). Regarding the presence of virulence genes, there was a significant increase in the presence of the cnf1 gene (P = 0.001) with early CAUTIs (3 - 6 days and 7 - 10 days) (Table 4).
| Demographic, Clinical Data, Biofilm Formation, and the Presence of Virulence Genes | Catheter Days 3 - 6 (n = 93) | Catheter Days 7 - 10 (n = 99) | Catheter Days > 10 (n = 12) | P-Value |
|---|---|---|---|---|
| Sex | 0.13 | |||
| Male | 54 (58.1) | 45 (45.5) | 8 (66.6) | |
| Female | 39 (41.9) | 54 (54.5) | 4 (33.3) | |
| Diabetes mellitus | 68 (73.1) | 58 (58.6) | 10 (83.3) | 0.046 |
| Kidney diseases | 72 (77.4) | 77 (77.8) | 7 (58.3) | 0.31 |
| Post-operative complications | 9 (9.7) | 10 (10.1) | 4 (33.3) | 0.045 |
| Fever | 54 (58.1) | 53 (53.5) | 4 (33.3) | 0.26 |
| Dysuria | 68 (73.1) | 75 (75.8) | 7 (58.3) | 0.43 |
| Urgency | 73 (78.5) | 73 (73.7) | 10 (83.3) | 0.63 |
| Frequency | 86 (92.5) | 87 (87.9) | 9 (75) | 0.16 |
| Pain on micturition | 41 (44.1) | 47 (47.5) | 7 (53.3) | 0.63 |
| Biofilm | 54 (58.1) | 49 (49.5) | 8 (66.7) | 0.33 |
| fim-H | 33 (35.5) | 27 (27,3) | 6 (50) | 0.19 |
| Pap-C | 10 (10.8) | 15 (15.2) | 4 (33.3) | 0.10 |
| Sfa | 12 (12.9) | 11 (11.1) | 2 (16.7) | 0.83 |
| Afa1 | 10 (10.8) | 13 (13.1) | 0 0 | 0.34 |
| Cnf1 | 2 (2.2) | 3 (3.03) | 3 (25) | 0.001 |
| Hyl | 3 (3.2) | 2 (2.02) | 0 0 | 0.73 |
a Values are expressed as No. (%).
5. Discussion
Catheter-associated urinary tract infections represent a major health problem worldwide. Many risk factors lead to CAUTIs despite the efforts of infection control guidelines (25). In the present study, the common comorbidities associated with CAUTIs were DM, renal disorders, and previous surgery history. There was a significant association between the early presence of CAUTIs (from 3 to 6 days) and the presence of DM (P = 0.046) and postoperative complications (P = 0.046). It is well known that DM predisposes individuals to various infections due to the compromised immune system (26). Different factors increase infections in diabetic patients, such as high glucose levels, which are considered a source of nutrient material for the bacteria, decreased T cell-mediated immunity, and impaired emptying of the bladder due to autonomic nervous system neuropathy (27).
Catheter-associated urinary tract infection as a complication of surgery is known due to the insertion of catheters for long durations in various surgical operations (28). Renal disorders, such as chronic renal failure, can be associated with increased urinary tract infections, especially in catheterized patients, due to a deficiency in the immune response and other comorbidities such as urinary obstruction and the presence of DM (27).
The early removal of the catheter is a cornerstone of the prevention of CAUTIs (29). However, the duration in which catheterization may lead to infections has not been fully studied. In the present study, urinary tract infections were commonly detected after 7 - 10 days of catheter insertion (48.53%), and then after 3 - 6 days of insertion (45.59%). A previous study reported a non-linear increase in the cumulative risk hazard as the increase in time of catheter insertion suggests that everyday increases in catheterization lead to an increased risk of developing CAUTIs (25). The present study provides interesting data about antimicrobial resistance models, biofilm production, and virulence genes associated with E. coli causing CAUTIs.
The capacity to form biofilm in an in vitro study was detected among 111 (54.4%) of isolated E. coli. The processes that predispose biofilm-bound E. coli to progress to CAUTI remain unclear but are of diagnostic and therapeutic interest (30). These processes are presumably complex and include biofilm efflux, adhesion to host tissue, evasion of the immune system, and nutrient acquisition (31). The capacity of organisms to form biofilm in an in vitro study indicates their capacity to form biofilm on the surface of the catheter and the tissues of the renal system, a finding that alerts clinicians that this biofilm is part of the pathogenesis of CAUTIs (32). The most frequently detected resistances to antibiotics among isolated E. coliwere toward ciprofloxacin, followed by resistance to the third-generation cephalosporins ceftazidime and cefotaxime, and resistance to trimethoprim/sulfamethoxazole.
The common antibiotics used for the treatment of UTIs include quinolones, cephalosporins, and trimethoprim-sulfamethoxazole. The unwise use of these antibiotics may explain the high frequency of resistance among isolated E. coliin the present study. The high resistance of the isolated E. colifrom CAUTIs to ciprofloxacin may be attributed to the presence of plasmids that encode resistance to this antibiotic, leading to the endemicity of these isolates in hospitals associated with hospital outbreaks of UTIs. The most frequently detected genes were fim-H (32.4%), followed by Pap-C (14.2%), SFA (12.3%), and afa1 (11.3%). The least frequently detected genes were cnf1 (3.9%) and hyl (2.5%). Higher rates of virulence genes were reported previously in E. coliisolated from UTI for Pap-C, hyl, and cnf1 (50.4%, 50.4%, 50.4% respectively) and lower frequency for afa1 (8.13%) (33). In another study, the prevalence rates for virulence genes sfa, pap, cnf1, afa, and hyl1 were 26%, 25%, 18.6%, 6%, and 5% respectively (34).
The high prevalence of the type 1 fimbria gene was in line with a previous study indicating a high incidence of type 1 fimbriae among uropathogenic E. coli(35), especially in CAUTIs. The difference in the prevalence of virulence genes among different studies can be attributed to the difference in virulence gene distribution among different strains of E. coli. There is also a known ability of E. colito exchange its genetic material and transfer genetic virulence factors, besides the effects of various antibiotics intake that leads to the elimination of certain strains with different virulence genes. Moreover, the presence of some virulence genes such as pap-c may be associated with certain types of UTIs, such as pyelonephritis, more than with its presence in cystitis (35). There was a significant association between the presence of fim-H, pap, afa-1, and hyl genes (P = 0.001, P = 0.001, P = 0.04, P = 0.046 respectively) and the capacity to form biofilm in isolated E. coli.
In another similar study by Zamani and Salehzadeh , it was found that biofilm-producing UPEC were significantly associated with the fim gene (36). The association of these adhesion genes and biofilm formation may indicate that these factors play a role in biofilm formation. Regarding the presence of virulence genes, there was a significant increase in the presence of the cnf1 gene and the early CAUTI insertion (3 - 6 days and 7 - 10 days). This finding may indicate that these adhesion genes play a role in the pathogenesis of CAUTIs shortly after catheter insertion.
The study’s focus on biofilm formation and virulence genes in CAUTIs represents a novel approach. Prior research has primarily examined bacterial colonization and antibiotic resistance, but this study delves deeper into the mechanisms driving infection persistence. This study is important for understanding biofilm dynamics, which is crucial for preventing and managing CAUTIs. Identifying specific virulence genes in CAUTIs can inform targeted therapies and also shed light on the genetic diversity of uropathogens. Finally, novel insights into biofilm and virulence genes can lead to more effective treatment strategies. We can implicate these findings in:
1. Catheter care guidelines: Healthcare policies should emphasize proper catheter insertion, maintenance, and removal.
2. antibiotic stewardship: Understanding virulence genes can guide antibiotic selection.
3. Biofilm disruption: Develop interventions to prevent or disrupt biofilm formation.
4. Gene-Based diagnostics: Incorporate virulence gene detection into diagnostic protocols.
5. Targeted therapies: Develop drugs that specifically target biofilm-associated virulence factors.
This study’s findings provide a fresh perspective on CAUTIs, emphasizing biofilm dynamics and virulence genes. By integrating these insights into healthcare policies and practice guidelines, we can enhance patient outcomes and reduce the burden of CAUTIs.
5.1. Conclusions
The present study highlights several risk factors leading to healthcare-associated urinary tract infections, including DM, renal disorders, and surgical intervention. Catheter-associated urinary tract infections usually occur within the first 10 days of catheter insertion. The presence of biofilm formation is a common finding with E. coli associated with CAUTIs. The virulence factors are common among isolated E. coli and are significantly associated with biofilm formation.
These findings can guide several aspects of healthcare policies, such as catheter care guidelines, which should emphasize proper catheter insertion, maintenance, and removal. Additionally, in antibiotic stewardship, understanding virulence genes can guide antibiotic selection and aid in developing interventions to prevent or disrupt biofilm formation. Finally, there is potential for developing drugs that specifically target biofilm-associated virulence factors. Further investigations are needed to identify novel targets for disrupting biofilms and enhancing antibiotic efficacy.