1. Background
Salmonellosis, a disease caused by Salmonella, represents a significant public health problem and economic burden worldwide (1-3). The transmission routes of Salmonella to humans are diverse, including consumption of contaminated food or water and contact with infected animals and humans. According to estimates from the World Health Organization (WHO), more than 90 million cases of human salmonellosis occur globally each year (4). Therefore, the control and prevention of salmonellosis are crucial.
The pathogenicity of Salmonella serovars depends on the presence of a variety of virulence genes, such as ssaR, spvC, pefA, sipA, fimA, sifA, sopE2, sopB, prgH, and stn. The ssaR and sifA genes are essential for the intracellular survival and subsequent replication of Salmonella (5-7), while the pefA, sipA, fimA, sopE2, sopB, and prgH genes are involved in adhesion and invasion of host cells (7-10). The spvC gene can suppress the innate immunity of infected hosts (11), and the stn gene is implicated in the complex mechanisms of diarrhea (12). Additionally, virulence genes can be transferred from one strain to another, enhancing the pathogenicity of Salmonella strains and potentially leading to disease outbreaks. For example, during the mid to late 1980s, there was an outbreak of typhoid fever in China characterized by severe symptoms and high mortality rates.
Subsequent studies identified the main causative agent of the epidemic as Salmonellaenterica serovar typhi (S. typhi) strains carrying the Salmonella plasmid virulence (spv) gene, which is typically absent in S. typhi (13). Investigating the presence of virulence genes in Salmonella strains is crucial for identifying pathogenicity, understanding potential transfer mechanisms, and assessing the risks posed by these strains. Thus, developing an efficient method for detecting Salmonella virulence genes is necessary for disease prevention and control.
PCR, including conventional and fluorescence PCR, is one of the most commonly used techniques for gene analysis. Fluorescence PCR introduces a fluorescent reporter (dsDNA binding dye or fluorophore-labeled probe) that binds to the amplification product and indicates its presence by fluorescence, allowing for real-time detection of the amplification product during the PCR reaction. The entire fluorescence PCR process can be completed in a closed-tube setting without any manual operation. Compared to conventional PCR, fluorescence PCR is simpler, faster, and easily automated for high-throughput testing (14). However, reports on the application of fluorescence PCR for analyzing Salmonella virulence genes are limited.
2. Objectives
The aim of this study is to develop a multiplex fluorescence PCR for detecting ten major virulence genes (ssaR, spvC, pefA, sipA, fimA, sifA, sopE2, sopB, prgH and stn) in Salmonella.
3. Methods
3.1. Strains
A total of 60 Salmonella strains, 2 Vibrio parahaemolyticus strains, and 3 Escherichia coli strains were utilized in this study. One S. typhimurium strain and two Salmonella London strains were isolated from meat, while other strains were isolated from patients with diarrhea. All strains were isolated and confirmed by biochemical characterization according to GB 4789 (national standards for microbiological examination of food in the People's Republic of China) and WS 271 (health standards for the diagnosis of infectious diarrhea in the People's Republic of China). Five Salmonella strains, comprising 3 strains of S. enteritidisand 2 strains of S. typhimurium (Table 1), were chosen as positive controls. Additionally, two V. parahaemolyticus and three E. coli strains served as negative controls. The virulence gene profiles of these control strains were detected by previously reported conventional PCR (15-20). Genomic DNA from all strains was extracted using the KAPA Express extract kit, following the manufacturer’s instructions.
| Isolates | Isolates No. | ssaR | spvC | pefA | sipA | fimA | sifA | sopE2 | sopB | prgH | stn |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Salmonella enteritidis | 1 | + | - | - | + | + | + | + | + | + | + |
| 1 | + | - | - | + | + | - | + | + | + | + | |
| 1 | + | + | - | + | + | - | + | + | + | + | |
| Salmonella typhimurium | 1 | + | + | + | + | + | + | + | + | + | + |
| 1 | + | - | - | + | + | + | + | + | + | + | |
| Vibrio parahaemolyticus | 2 | - | - | - | - | - | - | - | - | - | - |
| Escherichia coli | 3 | - | - | - | - | - | - | - | - | - | - |
3.2. Oligonucleotide Primers
The DNA sequences of ten major virulence genes in Salmonella were downloaded from GenBank and aligned using Clustal X 1.8. Based on this sequence alignment, relatively conserved regions were selected for primer design using Primer Premier 5.0. The details of the ten primer pairs targeting these genes are shown in Table 2.
| Variables | Target Gene | Primer | Sequence |
|---|---|---|---|
| Tube1 | ssaR | ssaR-F | 5’- CAAAAAAACTCTGAAGAGAAGGAAGCC-3’ |
| ssaR-R | 5’- GCCTGCGTTAACTGACTCACCGT-3’ | ||
| spvC | spvC-F | 5’- TGGGGCGGAAATACCATCTACAAAT-3’ | |
| spvC-R | 5’- CTGAATAGTCAGGCACATCCAGCG-3’ | ||
| pefA | pefA-F | 5’- ACCAGCCGGGTAATTTTGTTGACT-3’ | |
| pefA-R | 5’- CAGCAGAAGCCCAGGAAACAGTG-3’ | ||
| sipA | sipA-F | 5’- CACGTCAGAAAAGGGCACTACGG-3’ | |
| sipA-R | 5’- CTCGACGCCAGCCAGTTGACTG-3’ | ||
| fimA | fimA-F | 5’-TAATGACCTCTACTATTGCGAGTCTGATGTT-3’ | |
| fimA-R | 5’- AGAAAGCCACGGCAGCGTTG-3’ | ||
| Tube2 | sifA | sifA-F | 5’- TTAAAAAGTGAAATCCTTACCAACTCCCC-3’ |
| sifA-R | 5’- TCCGCTTTTGCTTTGCCAGTAGAA-3’ | ||
| sopE2 | sopE2-F | 5’- TAACATAACACTATCCACCCAGCACTACAG-3’ | |
| sopE2-R | 5’- TAAAACGATCTGACAGACTCGTCGACA-3’ | ||
| sopB | sopB-F | 5’- ATAAAGATGGCGATCTACAGACGGTAAA-3’ | |
| sopB-R | 5’- CCCACATTAAATGCGGCGACG-3’ | ||
| prgH | prgH-F | 5’- CGGCTGTGAGTTTCCATTGCTG-3’ | |
| prgH-R | 5’- TTCTAACTTCTCAGGCTGCTCGGG-3’ | ||
| stn | stn-F | 5’- CGCCAGCCCTTTGATGGACG-3’ | |
| stn-R | 5’- GGCATCAGCGTTATCAGCGCTC-3’ |
3.3. Optimization of Multiplex Fluorescence PCR
Primer pairs for ten virulence genes were distributed across two reaction tubes, with the distribution of these pairs detailed in Table 2. The multiplex fluorescence PCR was optimized by adjusting one parameter at a time. The parameters evaluated included the target primer concentration, which ranged from 0.05 µM to 1 µM, and the annealing temperature, which varied from 55 °C to 65 °C.
The multiplex fluorescence PCR was conducted using a 20 µL reaction mixture for each tube, consisting of 1 µL of DNA template, 1 µL of EvaGreen dye, 10 µL of KAPA 2G Fast Multiplex PCR Mix, appropriate amounts of target primers, and the remaining volume filled with distilled water. The PCR amplification took place in a Bio-Rad CFX 96TM real-time PCR system with the following program: An initial denaturation at 95 °C for 3 minutes, followed by 30 cycles of denaturation at 95 °C for 15 seconds, annealing for 30 seconds at the specified annealing temperature, and extension at 72 °C for 10 seconds.
3.4. Melting Temperature Curve Analysis
Fluorescence melting temperature curve analysis was conducted following PCR amplification. The PCR products were initially cooled to 78 °C and subsequently heated to 91 °C at a rate of 0.1 °C per second. Fluorescence signals were continuously monitored throughout the melting temperature curve analysis process.
4. Results
4.1. Optimization of Multiplex Fluorescence PCR
To develop a multiplex fluorescence PCR capable of detecting ten primary virulence genes in Salmonella, a systematic study was conducted to optimize the conditions. The optimal primer concentrations were determined as follows: In tube 1, 0.4 µM for ssaR, 0.4 µM for spvC, 0.3 µM for pefA, 0.1 µM for sipA, and 0.1 µM for fimA; in tube 2, 0.4 µM for sifA, 0.2 µM for sopE2, 0.2 µM for sopB, 0.05 µM for prgH, and 0.1 µM for stn. The optimal annealing temperature was established at 60°C.
4.2. Melting Temperature Curve Analysis of PCR Products
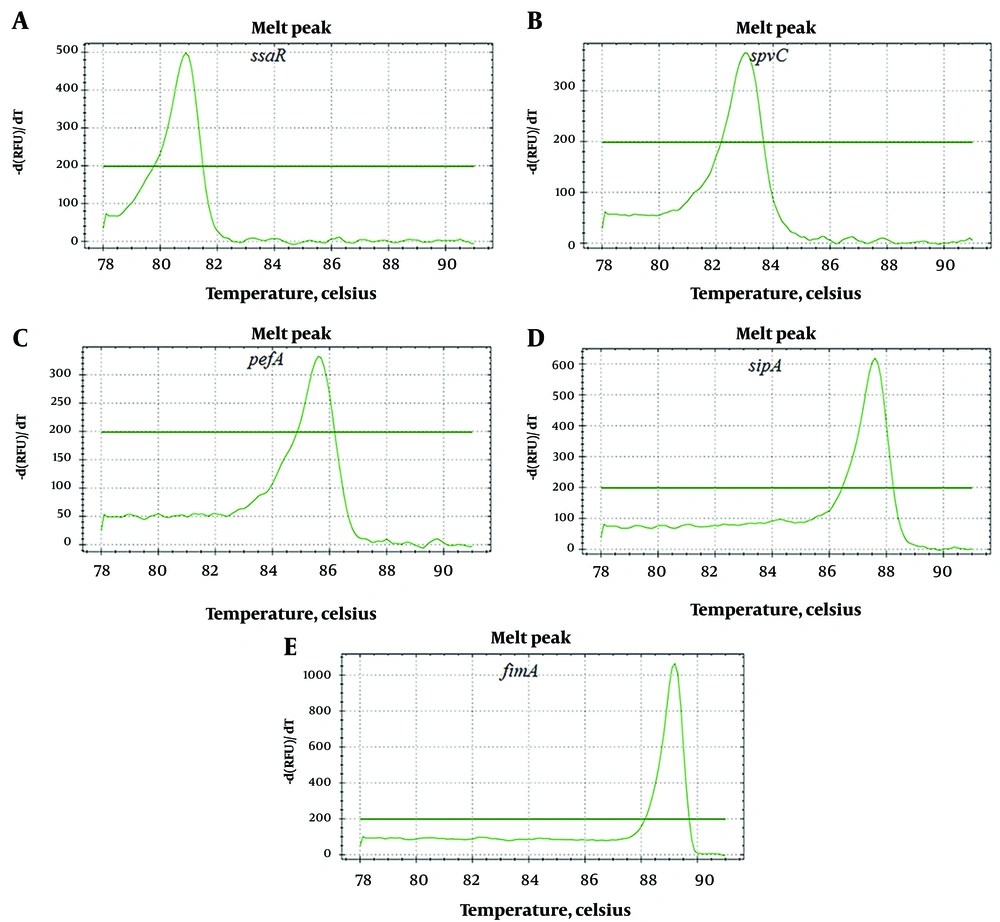
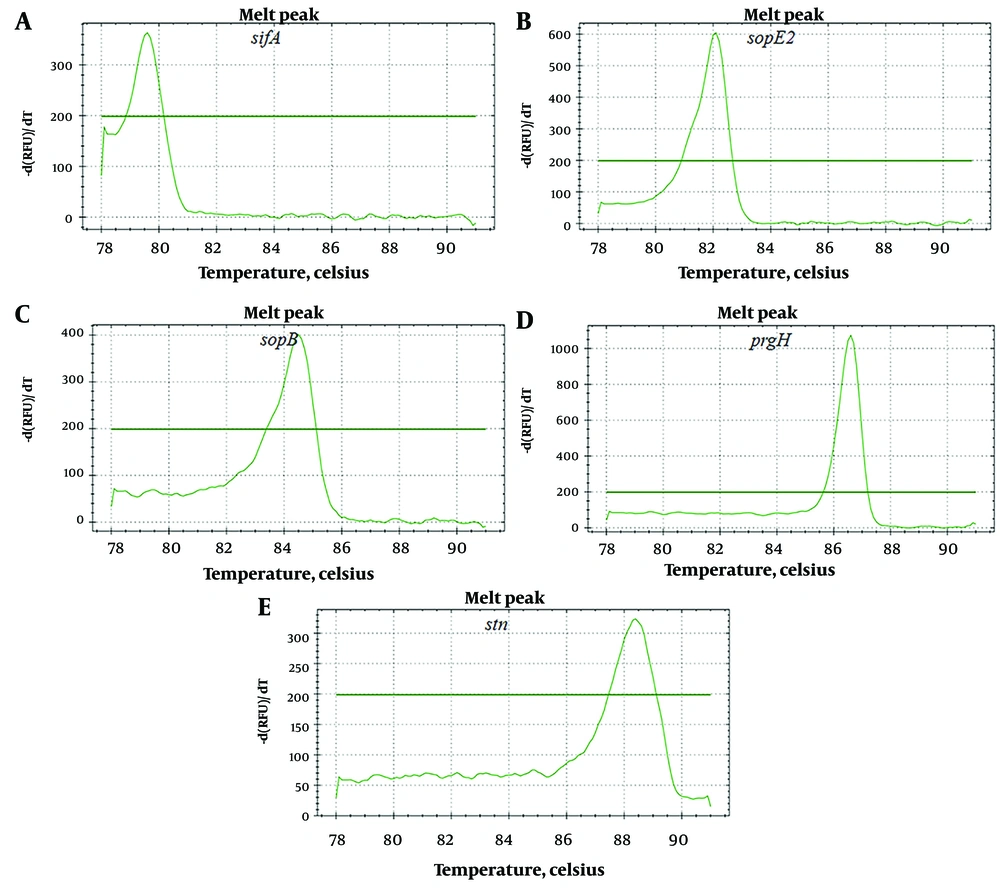
After PCR amplification and melting temperature curve analysis, ten distinct melting peaks were observed. The melting peak temperatures for the PCR products were as follows: ssaR primer pairs at 80.7 ± 0.3°C, spvC primer pairs at 82.8 ± 0.2°C, pefA primer pairs at 85.5 ± 0.1°C, sipA primer pairs at 87.4 ± 0.2°C, and fimA primer pairs at 89.0 ± 0.2°C (Figure 1). Additionally, the melting peak temperatures for sifA primer pairs were at 79.6 ± 0.2°C, sopE2 primer pairs at 81.8 ± 0.3°C, sopB primer pairs at 84.3 ± 0.2°C, prgH primer pairs at 86.3 ± 0.1°C, and stn primer pairs at 88.1 ± 0.3 °C (Figure 2).
4.3. Evaluation of Specificity of Multiplex Fluorescence PCR
Analysis of five positive controls and five negative controls using the newly developed multiplex fluorescence PCR showed that the five positive controls produced specific melting peaks, whereas no obvious melting peaks were observed in the five negative controls. The virulence genes profiles in the five positive controls detected by the newly developed multiplex fluorescence PCR were consistent with our previous results achieved by conventional PCR.
4.4. Analysis of Salmonella Strains
The distribution of 10 virulence genes in 60 Salmonella strains is summarized in Table 3. All strains contained seven or more of the tested virulence genes. The positive rates for virulence genes ssaR, sipA, sopE2, sopB, prgH, and stn were 100%, while sifA was 81.67%. However, the positive rates for spvC and pefA were relatively low, at 5% and 3.33% respectively. These results were confirmed by previously reported conventional PCR (15-20).
| Isolates | Isolates No. | ssaR | spvC | pefA | sipA | fimA | sifA | sopE2 | sopB | prgH | stn |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Salmonella enteritidis | 11 | + | - | - | + | + | + | + | + | + | + |
| Salmonella enteritidis | 5 | + | - | - | + | + | - | + | + | + | + |
| Salmonella enteritidis | 1 | + | + | - | + | + | - | + | + | + | + |
| Salmonella typhimurium | 1 | + | + | + | + | + | + | + | + | + | + |
| Salmonella typhimurium | 8 | + | - | _ | + | + | + | + | + | + | + |
| Salmonella stanley | 7 | + | - | _ | + | + | + | + | + | + | + |
| Salmonella agona | 3 | + | - | _ | + | + | - | + | + | + | + |
| Salmonella london | 5 | + | - | _ | + | + | + | + | + | + | + |
| Salmonella london | 1 | + | - | + | + | - | + | + | + | + | |
| Salmonella essen | 3 | + | - | - | + | + | + | + | + | + | + |
| Salmonella give | 1 | + | + | + | + | + | + | + | + | + | + |
| Salmonella tshiongwe | 1 | + | - | - | + | + | - | + | + | + | + |
| Salmonella montevideo | 2 | + | - | - | + | + | + | + | + | + | + |
| Salmonella abony | 3 | + | - | - | + | + | + | + | + | + | + |
| Salmonella weltevreden | 5 | + | - | - | + | + | + | + | + | + | + |
| Salmonella dublin | 3 | + | - | - | + | + | + | + | + | + | + |
5. Discussion
Infection with Salmonella is still one of the most important public health problems in the world. It can cause diseases ranging from gastroenteritis to typhoid fever (21). The pathogenicity of Salmonella strains is determined by their abilities to adhere, invade and replicate inside host cells, as well as to escape from or neutralize the host’s defenses (7, 22). These abilities of Salmonella strains depend on the virulence genes that they possess. In many cases, when a Salmonella serotype acquires new virulence genes that were previously absent, it can enhance pathogenicity of Salmonella strains and cause disease outbreak (13). Therefore, analysis of virulence genes profiles of Salmonella strains is necessary for preventing outbreak of disease caused by Salmonella infection. As one of the most widely used techniques in genes analysis, several PCR methods have been developed for Salmonella virulence genes analysis (15-20). However, these reported PCR methods for Salmonella virulence genes analysis are conventional PCR.
The drawbacks of conventional PCR are obvious. First, post-PCR analysis depend on gel electrophoresis is necessary for conventional PCR. The procedure of post-PCR analysis is complex and labor-intensive, which results in low throughput of the conventional PCR. Second, conventional PCR has the relative high risk of cross contamination as the essential step of opening PCR tube for post-PCR analysis. Therefore, other method have been developed for Salmonella virulence genes analysis. Though these methods have their advantages, they can’t completely replace PCR in the field of Salmonella virulence genes analysis. For example, microarray has a higher throughput than conventional PCR in Salmonella virulence genes analysis, PCR is still needed to validate the microarray data demonstrating weaker signals.
In this study, we developed a multiplex fluorescence PCR for Salmonella virulence genes analysis. Fluorescence PCR differs from the traditional PCR. The whole detection process of fluorescence PCR could be completed in a closed-tube setting and the post-PCR analysis depend on gel electrophoresis is not required.Without post-PCR analysis, fluorescence PCR increases assay throughput and reduces the risk of cross contamination (23). Thus, our newly developed multiplex fluorescence PCR for Salmonella virulence genes analysis overcomes the drawbacks of conventional PCR. Our multiplex fluorescence PCR encompasses ten main virulence genes (ssaR, spvC, pefA, sipA, fimA, sifA, sopE2, sopB, prgH, and stn), which play crucial roles in the diseases caused by Salmonella. These genes are significant indicators of pathogenicity, facilitating the ability of Salmonella to adhere to, invade, and replicate inside host cells, and to escape from or neutralize the host's defenses (5, 12).
In this study, all analyzed strains contained seven or more of the tested virulence genes. The positive rates for virulence genes such as ssaR, sipA, sifA, sopE2, sopB, prgH, and stn were relatively high, whereas those for spvC and pefA were relatively low. The distribution of most virulence genes in our study aligns with results from previous reports. For instance, Yue et al. (15) analyzed 61 Salmonella isolates from the feces of children with acute diarrhea and found that all isolates (100%) tested positive for prgH, 51 isolates (83.61%) for sopB, and 11 isolates (18.03%) for pefA. Another study by Qiao et al. (24) reported prevalence rates for the virulence genes sipA, pefA, and spvC at 77.6%, 10.3%, and 1.9%, respectively.
5.1. Conclusions
In summary, the newly developed multiplex fluorescence PCR provides a straightforward, cost-effective, and high-throughput method for detecting virulence genes in Salmonella. Thus, it has the potential to become a routine method for analyzing Salmonella virulence genes.