1. Background
Antimicrobial resistance (AMR) has emerged as a global health concern, with both clinical and public environments acting as hotspots for the spread of antibiotic resistance. Water environments, in particular, are significant reservoirs for antibiotic-resistant bacteria and genes. Due to public gatherings, recreational freshwater and marine beaches face a wider range of potential health risks (1). In recent years, the rapid spread of multidrug-resistant Gram-negative bacteria, especially the increase in carbapenem-resistant Gram-negative bacteria, has led to critical levels of multidrug resistance in clinical settings. Colistin has been re-applied clinically and is considered the last line of defense against Gram-negative bacteria (2).
Researchers have found an increasing number of Gram-negative bacteria resistant to colistin in domesticated animals, the environment, and even humans (3). In 2015, a colistin resistance gene mediated by a horizontal transfer plasmid was discovered in porcine Escherichia coli plasmids (4, 5), which rapidly spread across more than 50 countries and regions worldwide (6). The epidemiological safety of recreational water has become a concern. Currently, there are few studies on the epidemic characteristics of antibiotic resistance based on the "One Health" approach in different media. From January to December 2019, our team conducted sampling at two bathing beaches, more than 30 swimming pools, and beaches in Hainan Province, as well as among practitioners.
2. Objectives
We aimed to determine the spatial distribution, genetic characteristics, and transmission of colistin-resistant bacteria in recreational freshwater and coastal beaches. In this study, after selecting strains, the antibiotic resistance and virulence characteristics of E. coli carrying the mcr-1 resistance gene were further studied through whole-genome resequencing. Multilocus sequence typing (MLST) and next-generation sequencing (NGS) data analysis were conducted with E. coli carrying the mcr-1 gene from China. The relationship of these strains to others from Asia, Europe, and the Americas was compared to construct a maximum likelihood evolutionary tree.
3. Methods
3.1. Source of Strains and Rationality
A total of 136 E. coli isolates were collected from swimming pool water and pool walls, seawater and sand samples, and anal swabs from beach staff with informed consent. The human strains used in this study underwent ethical review by the affiliated Hainan Hospital, Hainan Medical University (2019075), and informed consent was obtained from all subjects involved. IBM SPSS 22.0 was used for statistical analysis.
3.2. Isolation, Culture and Identification of Single Strains
Processing of water samples: Samples were randomly collected. 10 mL of water samples were loaded into aseptic centrifuge tubes, stored in refrigerated containers, and processed within 4 hours. The tubes were centrifuged at 3000 rpm for 5 minutes, and 200 μL of the precipitate was saved on MacConkey Agar (MAC). The precipitate was then transferred to each MAC plate and evenly spread using an inoculation ring. A sterile cotton swab was used to wipe a 5 cm × 5 cm area of the wet swimming pool wall. Following aseptic procedures, 10 mL of distilled water was added, shaken for 1 minute to mix, and left for 1 hour before being inoculated using the aforementioned water sample processing method. Human anal swab samples were collected from the staff of the two bathing beaches and inoculated on MAC plates. All inoculated MAC plates were placed in a 35°C incubator for 48 hours, with colony growth observed every 24 hours. Suspicious pink colonies were selected for MALDI-TOF MS identification and subculture, and then stored at -80°C.
3.3. Materials and Equipment
Mass spectrometer VITEK MS (France Biomerieux), polymyxin B drug sensitivity reagent (Kangtai Bio), Vitek2 automatic bacterial identification instrument (France Biomerieux), MacConkey medium (China Guangdong Huankai), serologic diagnostic serum for E. coli O157:H7 (Ningbo Tianrun, China), Tanon™ nucleic acid dye (China Shanghai Tianneng), 2 × Taq PCR master mix (Beijing Tiangen, China), Gradient PCR instrument (Eppendorf, Germany), automatic digital gel image analysis system (Tanon-4100, Tianneng, China), constant temperature shaking table (Shanghai Zhichu Instrument Co., Ltd.), and prokaryotic genome NGS sequencing were sent to Megi Biological Company.
3.4. 16S Sanger Sequencing Identification, Antibiotic Resistance and Virulence Experiment
The 16S Sanger identification method was used to identify the strains carrying mcr-1. The colistin (PB) sensitivity test was performed using the microbroth dilution method, with ATCC25922 as the quality control strain. The minimum inhibitory concentration (MIC) value interpretation standard was determined by referencing the critical values of sensitivity (S ≤ 2 mg/L) and resistance (R > 2 mg/L) set by the European Committee for Antimicrobial Susceptibility Testing (EUCAST) in 2020 (7). For other antibiotic susceptibility tests, the Vitek2 Compact automatic bacterial identification instrument from Biomerieux and the N334 card were used, with MIC values determined according to the American Clinical and Laboratory Standards Institute (CLSI) guidelines in 2019 (8). The quality control strains used were E. coli ATCC25922 and Pseudomonas aeruginosa ATCC27853.
For the O157:H7 serum slide agglutination test, one drop of O157:H7 serum was added to a clean slide. A small amount of the tested bacteria was then mixed with the serum, and the slide was gently shaken. The result was visually assessed within 1 minute, using physiological sodium chloride solution as a negative control. The results were interpreted as follows: Obvious agglutination within 1 minute was considered positive, while uniform turbidity was considered negative.
3.5. Extraction of Bacterial DNA
DNA extraction was performed according to the kit instructions. The supernatant of 70 - 80 μL was transferred to a new EP tube to obtain the DNA, and the optical density (OD) value was measured. The OD260/280 ratio ranged from 1.8 to 2.1.
3.6. PCR Amplification and Sequencing
The primers for the mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 genes were synthesized by Shanghai Bioengineering Company (9). The reaction conditions were as follows: Pre-denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 15 s, annealing at 58°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 5 min. After amplification, the product was subjected to electrophoresis on a 1.5% agarose gel at 120V for 40 minutes. The amplified product was then sent to Shanghai Shenggong Bioengineering Company for sequencing. Strains were identified using 16S Sanger sequencing with primer sequences 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT). The target genes, specifically the virulence genes stx and rfbE, were amplified using PCR (10).
3.7. Prokaryotic Genome Next-Generation Sequencing (NGS) Sequencing
The four strains were sent to Megi Bio-Company for genome NGS sequencing. SPAdes nucleic acid splicing software was used to assemble the FASTQ sequencing files that passed quality testing through the denovo non-reference genome assembly mode. The gene assembly quality report was generated using Quast software. The genome sketch was assembled using the reference strain K12_MG1655 (4641652 bps) genome. Based on the assembled genome sketch and scaffolds, drug resistance genes and virulence genes were identified using the CARD and VFD databases.
3.8. Mobile Components Analysis
The prediction software for insertable sequences was ISEScan_v1.7.2.1. The detection software for integrons and clustered regularly interspaced short palindromic repeats (CRISPR) were Integron_Finder_v2.0 and CRISPR Finder, respectively. Mobile elements were used to analyze foreign genomic fragments in the genome.
3.9. Multilocus Sequence Typing (MLST) Genotype
Multilocus sequence typing was performed using the Institute Pasteur typing method (11), amplifying and sequencing eight steward genes synthesized by BGI Genomics Co., Ltd: dinB, icdA, pabB, polB, putP, trpA, trpB, and uidA (Appendix 1). The resultant gene sequences for each strain were compared with the reference sequences stored on the Institute Pasteur website. Eight allele genes were obtained through this comparison, which were used to determine the sequence type (ST) of the strain.
3.10. Comparative Analysis of Pangenomic Isolates with Characteristic Genes
The NGS data of 80 E. coli strains were selected from the National Center for Biotechnology Information (NCBI) in January 2020. The mcr-1 gene of E. coli was used with the GenBank DNA sequence MG763104.1d serving as a reference. Comparative analysis was conducted using the basic local alignment search tool (BLAST). Gene annotation was performed using Prokka software. Pan-genomic analysis of protein sequences was conducted using bacteria pan genome analysis (BPGA) software, which identified the core genes, auxiliary genes, unique genes, and deletion genes of all strains (12). The phylogenetic tree of core genes was constructed based on these core genes, and an additional phylogenetic tree was constructed using all identified genes.
4. Results
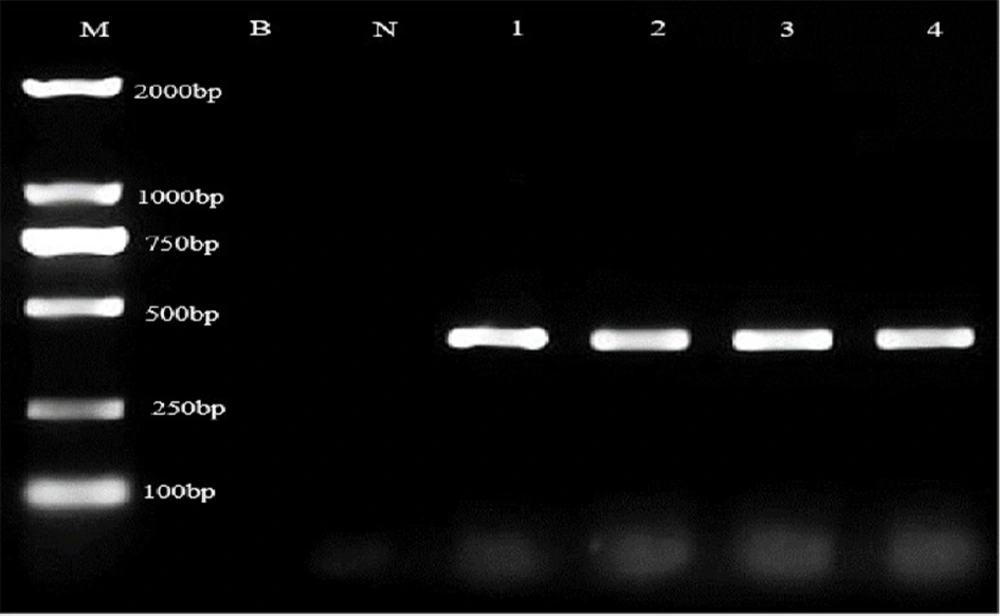
A total of 136 E. coli strains were screened in this study, among which 4 mcr-1 target strains showed bands at 320 bp during PCR detection (Figure 1), accounting for 2.94% (4/136). The positive rates of mcr-1 strains from the swimming pool, bathing seawater, and anal swab samples were 2/40, 1/28, and 1/68, respectively. The statistical difference between the different sources was not significant (χ2 = 1.148, P>0.05). The corresponding strain codes were A93, A135, AD66, and ADPW66. No other mcr family genes were found.
4.1. Identification and Drug-Resistant Phenotypes of Escherichia coli
The sequencing results of 16S rRNA genes were compared with the MW082015.1 sequence in GenBank using BLAST, and the results showed 100% homology. Four target strains carrying the mcr-1 resistance gene were re-checked by 16S rRNA sequencing. The sequence showed 100% homology with the GenBank number CP100005.1, and the strains were identified as E. coli. The drug susceptibility test results were obtained using the microbroth dilution method combined with the Vitek2 Compact automatic bacterial identification instrument from France Biomerieux and the matching N334 card (Table 1).
| Antibiotics(mg/L) | NO. of Strain/Source of Strain | |||
|---|---|---|---|---|
| Sea Water | Human | Swimming Pool Well | Swimming Water | |
| No. 93 | 135A | ADPW66 | AD66 | |
| LEV | 1 | 1 | 1 | 1 |
| SXT | ≥ 8/152 | ≥ 8/152 | ≥ 8/152 | ≤ 0.5/9.5 |
| TGC | ≤ 4 | ≤ 4 | ≤ 4 | ≤ 4 |
| AMP | ≥ 32 | ≥ 32 | ≥ 32 | ≥ 32 |
| AMK | ≤ 4 | ≤ 4 | ≤ 4 | ≤ 4 |
| GEN | ≤ 1 | ≤ 1 | ≤ 1 | ≤ 1 |
| CHL | ≤ 8 | ≥ 32 | ≥ 32 | ≥ 32 |
| IPM | ≤ 1 | ≤ 1 | ≤ 1 | ≤ 1 |
| AMS | = 16/8 | ≤ 2/1 | ≤ 2/1 | ≤ 2/1 |
| TZP | ≤ 4/4 | ≤ 4/4 | ≤ 4/4 | ≤ 4/4 |
| CTX | ≤ 4 | ≤ 4 | ≤ 4 | ≤ 4 |
| CAZ | ≤ 4 | ≤ 4 | ≤ 4 | ≤ 4 |
| FEP | ≤ 2 | ≤ 2 | ≤ 2 | ≤ 2 |
| CXM | ≤ 8 | ≤ 8 | ≤ 8 | ≤ 8 |
| PB | 4 | 8 | 8 | 8 |
Abbreviations: R, drug resistance; I, intermediate; S, sensitive; LEV, levofloxacin; SXT, compound sulfamethoxazole; TGC, tigecycline; AMP, ampicillin; AMK, amikacin; GEN, gentamicin; CHL, chloramphenicol; IPM, imipenem; SAM, ampicillin/sulbactam; TZP, piperacillin/tazobactam; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; CXM, cefuroxime; PB, polymyxin.
4.2. Results of Prokaryotic Genome Next-Generation Sequencing (NGS) Sequencing
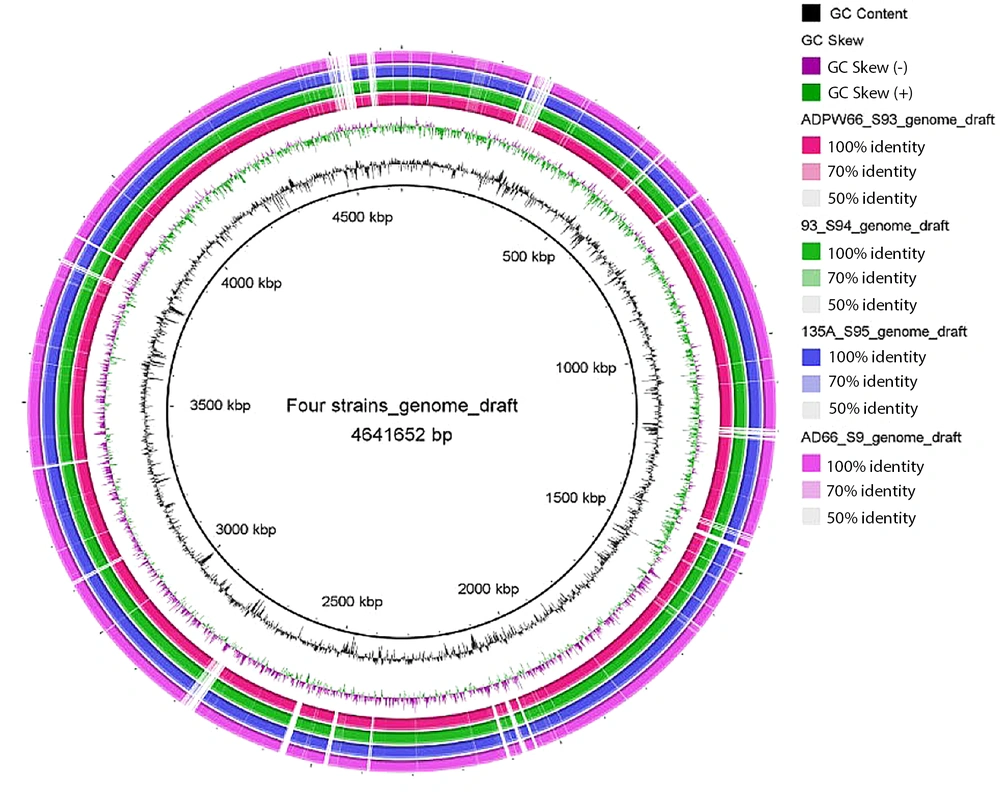
SPAdes nucleic acid splicing software was used to splice FASTQ sequencing files that met the quality detection criteria for 4 strains of E. coli with the mcr-1 resistance gene through de novo non-reference genome assembly mode. The draft genome was assembled using the reference K12_MG1655 (4641652 bps) genome (Figure 2).
Plot of the draft genome of the genome for four Escherichia coli strains carrying mcr-1. Note: The reference strain K12_MG1655 (black) Scale is shown in the innermost circle. The second circle shows the G + C content (deviation from the mean), and the third circle shows the G + C skew (green, +; Purple, -). The fourth, fifth, sixth, and seventh circles indicate that the genomes numbered ADPW66 (red), 93 (green), 135A (blue), and AD66 (purple) are compared with the genomes of the reference strain K12MG1655.2.3 mobile components and resistance gene analysis.
4.2.1. Mobile Elements of 4 Strains Carrying Mcr Gene
As shown in Table 2, the insertable sequence IS3 was the most widely distributed type. ISKpn26 and ISEhe3 were more prevalent in A135 and ADPW66. Strain A93, isolated from the marine beach, had the highest number of unknown insertion sequences, with five detected. Additionally, the strain from a human source exhibited the highest number of antibiotic resistance genes (ARGs).
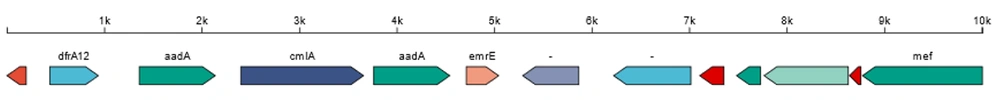
Mobile elements in the longest Scaffold52 for the isolate from marine water were identified. The human-sourced strain 135A has two categories of integrons. The longest integron is located in Scaffold52, which is 5425 bp in length and carries 6 ARGs (Figure 3). Strain 135A has unique ARGs including dfrA12, aadA2, sul3, and qacH. While mcr4.4 and mcr5.2 were identified in the further search for mcr family resistance genes, their identity was only below 40%. The following prevalent transmissible ARGs were identified: Mcr-1, QnrS1, tet(A), TEM-176, and CmlA1/floR. The CRISPR types of the four strains were classified as CRISPR1 and CRISPR2, but the number of different types varied. The A93 strain, isolated from the beach, exhibited a limited presence of CRISPR elements, as shown in Table 2.
| Strains Name | IS Name | IS Family | Length (bp) | CRISPR No. |
|---|---|---|---|---|
| 135A | ISEc1 | ISAs1 | 1684 | 18 |
| IS30D | IS30 | 1230 | ||
| ISKpn26 | IS5 | 1197 | ||
| AD66 | ISEhe3 | IS3 | 783 | 15 |
| IS2 | IS3 | 1071 | ||
| - | IS91 | 852 | 17 | |
| ADPW66 | ISCro3 | IS4 | 1033 | |
| ISKpn26 | IS5 | 1197 | ||
| A93 | - | ISNCY | 1153 | 13 |
| - | IS5 | 1144 | ||
| ISCro3 | IS4 | 1033 | ||
| ISEhe3 | IS3 | 783 | ||
| IS2 | IS3 | 1071 |
4.3. Genotyping Results
Two typing methods were performed for bacterial genomic characterization, comparing the results with those in the database. Multilocus sequence typing utilized allelic and genotypic information from over 100 species or genus-level bacteria available in the PubMLST database (https://pubmlst.org/ ,MLST2.0). The housekeeping genes dinB, icdA, pabB, polB, putP, trpA, trpB, and uidA8 were selected from the database. The ST type of three strains (135, AD66, ADPW66) was most similar to ST987, while strain No. 93 was most similar to ST24, with the allele profile shown in Table 3. The electrophoretogram of the eight housekeeping genes is shown in Appendix 2. All gene bands were between 400 - 700 bp, and the sequencing results were identical to the BLAST results.
| Sample | dinB | icdA | pabB | polB | putP | trpA | trpB | uidA | ST |
|---|---|---|---|---|---|---|---|---|---|
| 93 | 7 | 32 | 18 | 2 | 5 | 8 | 2 | 2 | 24 |
| 135A | 10 | 2 | 3 | 3 | 18 | 1 | 4 | 2 | 987 |
| AD66 | 10 | 2 | 3 | 3 | 18 | 1 | 4 | 2 | 987 |
| ADPW66 | 10 | 2 | 3 | 3 | 18 | 1 | 4 | 2 | 987 |
4.4. Virulence Gene Analysis
The virulence analysis showed that all the E. coli isolates carried the ibeC virulence gene, which belongs to E. coli O157:H7 (EHEC serotype). However, the O157 and H7 specific gene Stx was not found, and the O157 serologic agglutination test was negative. The cfaA, cfaB, cfaC, and cfaD/cfaE genes were found in all strains, which are associated with E. coli O78:H11:K80 H10407 (ETEC serotype). The virulence genes fimB, fimC, fimD, and fimE were also found in all the E. coli strains. According to the five types of E. coli causing diarrhea, the target gene (as per the Standard of GB 4789.6 2016, China) was identified. The virulence gene uidA was detected in all cases, but other associated genes were not positive.
4.5. Four Target Strains of Homology Analysis
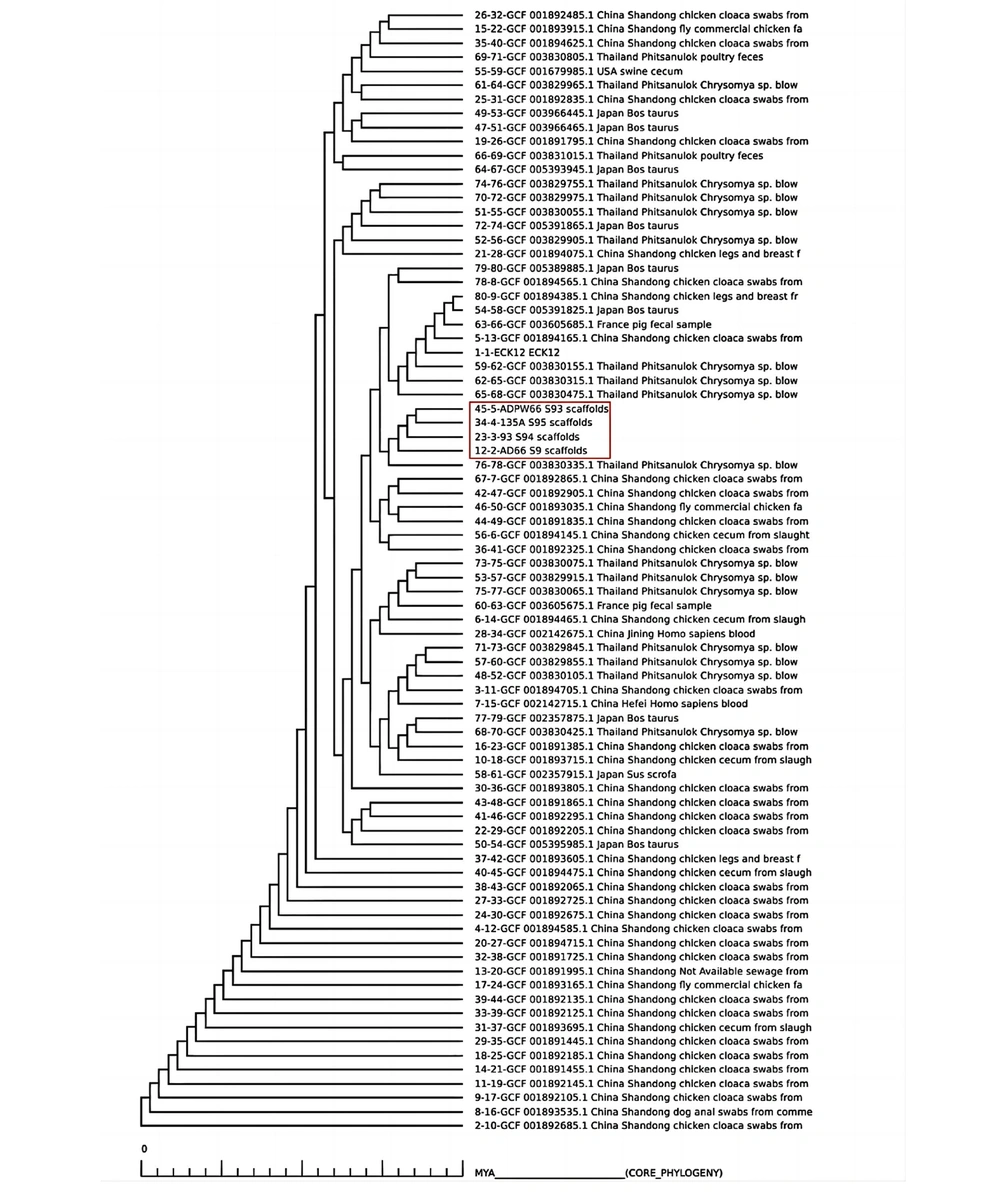
Four E. coli isolates carrying mcr-1 were compared and analyzed using pangenomic analysis. Next-generation sequencing data of E. coli from 2016 to 2019 were selected from the NCBI website, and BLAST was performed. A total of 80 other mcr-1 positive E. coli strains were obtained. The data were selected from representative regions worldwide, including the United States, Australia, Japan, Vietnam, France, and Thailand. Bacteria pan genome analysis software was used to analyze the protein sequences, obtaining the core genes, auxiliary genes, unique genes, and deletion genes of all strains. The phylogenetic tree of core genes was drawn.
In Figure 4, the closest genetic relative observed was Chrysomya sp. from Phitsanulok, Thailand. The second closest strains were obtained from chicken cloaca swabs in Shandong Province, China, but it lies outside the main branches. The two most closely related isolates were sourced from an anal swab and a swimming pool wall.
5. Discussion
The prevalence of drug-resistant strains can be transmitted through cloning in environments, animals, and humans. In turn, ARGs can cause local outbreaks or widespread dissemination of resistant bacteria through transformation, conjugation, and transduction (13, 14). Four colistin-resistant E. coli strains carrying the mcr-1 gene were isolated from artificial swimming pools, beaches, and related personnel in two important cities located in the south and north of Hainan Province. This finding suggests that mcr-1E. coli strains exist in close living environments of humans. The qnrS gene of these four strains did not show the phenotype of resistance to quinolones. Insertable sequences belonging to the IS5 and IS3 families were prevalent in the four strains, and human and freshwater strains also contained ISKpn26, which has been reported in selected Klebsiella pneumoniae strains (15, 16). The ST987 E. coli type harboring mcr-1 was found scattered in human feces, swimming water, and on the pool wall, revealing a widespread epidemic in tropical Hainan. The seawater isolate A93 showed a low level of resistance to colistin, while the other three target strains showed a moderate level of resistance to colistin.
The results of the prokaryotic genome sequencing were compared with the phylogenetic tree of the core genes of the mcr-1 positive E. coli isolates from China and abroad, indicating a close relationship between the mcr-1 carrying isolate from Chrysomya sp. in Thailand. This suggests that the transmission route involving travelers could not be excluded. Additionally, the mcr-1 positive strain also carried various ARGs, which led to its resistance to ampicillin, cotrimoxazole, and chloramphenicol. Phylogenetic analysis of the genome can be used to understand the transmission mode of pathogens or antibiotic-resistant bacteria. The use of public databases helps to trace the most likely source of transmission. Common methods for constructing phylogenetic trees include constructing them based on the core genome, pan-genome, single nucleotide polymorphism, gene copy number, and non-coding conserved genes. With the application of NGS technology, the core genome is usually combined with specific sequence characteristics and offers certain advantages (17).
5.1. Conclusions
Currently, the increasing prevalence of the mcr family genes is raising concerns due to its potential risks to global health. Particular attention should be paid to genes with strong migration abilities, such as mcr-9, and the changes in drug resistance (18). Enhancing the monitoring of bacteria in recreational freshwater and coastal environments is crucial for preventing the spread of antibiotic microbial resistance in tropical tourist areas. It is necessary to trace the sources of multi-environmental antibiotic-resistant bacteria in recreational bathing pools and beaches, which are in close contact with humans.