1. Background
The highly pathogenic severe acute respiratory syndrome-related coronavirus (SARS-CoV-2), which originated in China in December 2019, poses a significant global public health threat (1). Approximately 20% of infected individuals exhibit varying degrees of illness, necessitating hospitalization, with 5% requiring ventilatory support in an intensive care unit (2). The most severe manifestations of the disease are linked to systemic inflammation (3). The underlying mechanisms of COVID-19 are still being investigated, and further research is required to understand the factors and biomarkers contributing to the diverse progression of the disease (4). It has been observed that patients with severe illness exhibit an acute inflammatory response characterized by elevated levels of certain inflammatory markers such as C-reactive protein (CRP), ferritin, D-dimer, and the neutrophil-to-lymphocyte ratio. Additionally, heightened levels of specific inflammatory cytokines and chemokines have been noted in severely affected patients (5-8).
To comprehensively understand the pathogenesis of COVID-19, it is crucial to delineate the roles of cytokines and chemokines across the spectrum of disease severity in various populations, age groups, and individuals with and without comorbidities. Identifying prognostic biomarkers will enable the prioritization of hospital resources and personalized treatment approaches. Consequently, there is considerable interest in elucidating how severe acute respiratory syndrome disrupts normal antiviral immune responses and the associated biomarkers. IP-10 (interferon-gamma inducible protein-10) is secreted by some cells such as monocytes in response to interferon-gamma and has anti-tumor activity. Macrophage inflammatory protein 1-alpha (MIP1α) is one of the most important factors produced by macrophages and monocytes and is very important in immune responses against infection and inflammation. Interleukin-6 (IL-6) is one of the important interleukins of the body, secreted from white blood cells, and plays a role in inflammatory and immune responses. Interleukin-1 beta and interleukin-1 alpha are cytokines involved in the regulation of immune responses and inflammatory reactions (9, 10).
2. Objectives
Our study aims to identify circulating concentrations of interferon-gamma inducible protein-10 (IP-10), macrophage inflammatory protein 1-alpha (MIP1α), interleukin-6 (IL-6), and interleukin-1 beta (IL-1β) as chemokines and cytokines in severe COVID-19 cases compared to mild cases and individuals who have recovered from the disease.
3. Methods
3.1. Human Samples
Participants for this study were recruited from Masih Daneshvari Hospital and included individuals diagnosed with severe COVID-19, individuals diagnosed with mild COVID-19, and individuals who had previously recovered from COVID-19 (Table 1). This study was conducted before the availability of COVID-19 vaccinations. Each group comprised 40 patients. Confirmation of SARS-CoV-2 infection in all enrolled patients was established through viral PCR analysis of nasal and pharyngeal swab specimens obtained during the acute phase of the infection, following the guidelines set by the World Health Organization.
| Disease Severity | Specifications |
|---|---|
| Mild | Mild clinical sign (fever above 38℃, with or without cough, no dyspnea) no visible sign of pneumonia |
| Medium | Fever; Respiratory symptoms, the visible sign of pneumonia |
| Severe | Respiratory distress, respiratory rate (RR) more than 30 times per minute |
| pulse oximetry oxygen saturation level (SPO2) less than 93% at rest | |
| The arterial partial pressure of O2 and the fraction of inspired oxygen (PaO2/FiO2) ratio less than 300 mmHg |
3.2. qRT-PCR
Plasma samples were utilized to extract RNAs of MIP1-α and IP-10, employing the RNeasy Midi Kit (Qiagen, Cat No. 75144) following the manufacturer's instructions. Subsequently, cDNA synthesis was performed on the extracted RNAs using the Viva 2-step RT-PCR Kit (Cat No. RTPL12). To quantify the expression of MIP1-α and IP-10, real-time PCR was conducted using the CinnaGreen qPCR Mix, 2X (Cat No. MM2041), with RNA18S serving as the endogenous control for normalization, as per the manufacturer's guidelines. Gene expression profiles were normalized to RNA18S and calculated using the ΔΔCt (2−ΔΔCt) method. The primer sequences employed for the expression study can be found in Table 2.
| Parameters | IP-10 | MIP1-a | 18S rRNA |
|---|---|---|---|
| Forward primer | GGTGAGAAGAGATGTCTGAATCC | TGCATCACTTGCTGCTGACACG | GTAACCCGTTGAACCCCATT |
| Length | 23 | 22 | 20 |
| Reverse primer | GTCCATCCTTGGAAGCACTGCA | CAACCAGTCCATAGAAGAGG | CCATCCAATCGGTAGTAGCG |
| Length | 22 | 20 | 20 |
| Length of amplified fragment | 134 | 333 | 152 |
| Annealing temperature | 59 | 60 | 56 |
3.3. Enzyme-linked Immunosorbent Assay
As part of routine venipuncture, peripheral blood samples were collected from each participant within six days of hospital admission. Through centrifugation at 1000×g for 15 minutes at 4°C, serum samples were isolated and promptly stored at -80°C until further analysis. The levels of soluble IL-6 and IL-1β were assessed using enzyme-linked immunosorbent assay (ELISA), following the instructions provided by the respective manufacturers. The optical density (OD) readings of IL-6 and IL-1β were measured spectrophotometrically using a plate reader at wavelengths of 450 nm and 620 nm, respectively. The levels of IL-6 and IL-1β were determined by referencing a standard curve.
3.4. Statistical Analysis
SPSS 24 software was used for statistical analysis. Differences between groups were analyzed by one-way ANOVA test with Tukey's HSD post-hoc. The correlation between variables was analyzed with the Spearman Rank. The area under the receiver operating characteristics (ROC) curve (AUC) was estimated for patients. The optimal cut-off value was determined by Youden’s index. All data are presented as mean ± SD, with P values less than 0.05 considered significant.
4. Results
The study encompassed a total of 120 individuals diagnosed with COVID-19. These participants were categorized into three distinct groups based on the severity of their condition: Severe, mild, and recovered COVID-19. Importantly, no significant variations in age were observed among the different groups. The mean ages (SD) of patients with severe, mild, and recovered COVID-19 were 55.62 (8.22), 54.48 (7.94), and 55.32 (8.88) years, respectively. Statistical analysis revealed no significant difference in age between the groups (P = 0.41).
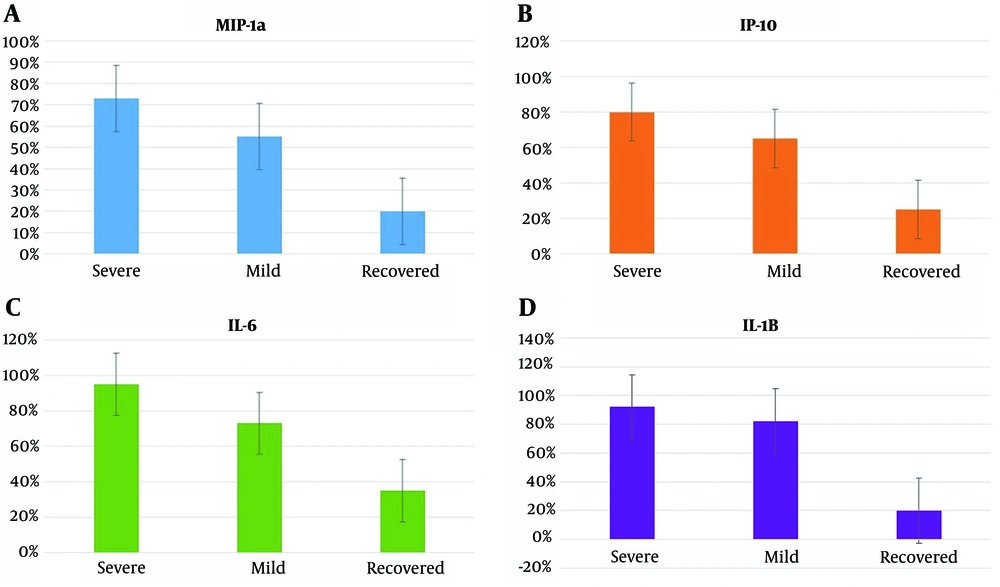
The biomarkers MIP1-α, IP-10, IL-6, and IL-1β exhibited positive results in varying percentages across the different infection groups. In the severe infection group, MIP1-α, IP-10, IL-6, and IL-1β were positive in 72.5%, 80%, 95%, and 92.5% of cases, respectively. Within the mild infection group, these biomarkers yielded positive results in 55%, 65%, 72.5%, and 82.5% of cases, respectively. For the recovered group, the percentages of positive results for MIP1-α, IP-10, IL-6, and IL-1β were 20%, 22%, 35%, and 20%, respectively, as depicted in Figure 1. Statistical analysis utilizing the two-sample binomial test was conducted to compare the positive rates of these biomarkers between the severe infection, mild infection, and recovered groups. The results revealed a statistically significant difference among the three groups under investigation (P < 0.001).
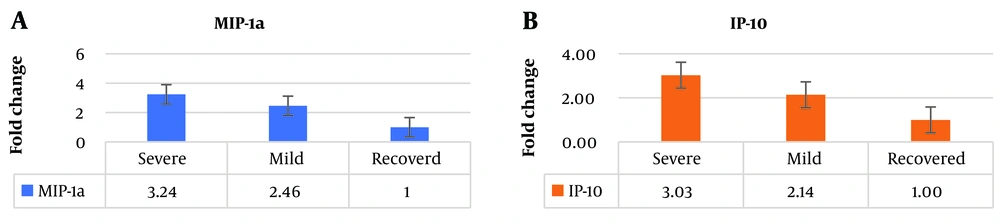
The expression level of MIP1-α was significantly higher in individuals with severe infection compared to healthy individuals, showing a 3.24-fold increase. Similarly, in individuals with mild infection, MIP1-α exhibited a 2.46-fold increase compared to healthy individuals. Furthermore, the expression level of IP-10 was 3.03 times higher in individuals with severe infection compared to healthy individuals. Likewise, in individuals with mild infection, the expression level of IP-10 was 2.14 times higher than in healthy individuals (Figure 2). These results indicate substantial differences in the expression levels of MIP1-α and IP-10 between different infection groups, suggesting their potential as valuable biomarkers for assessing infection severity.
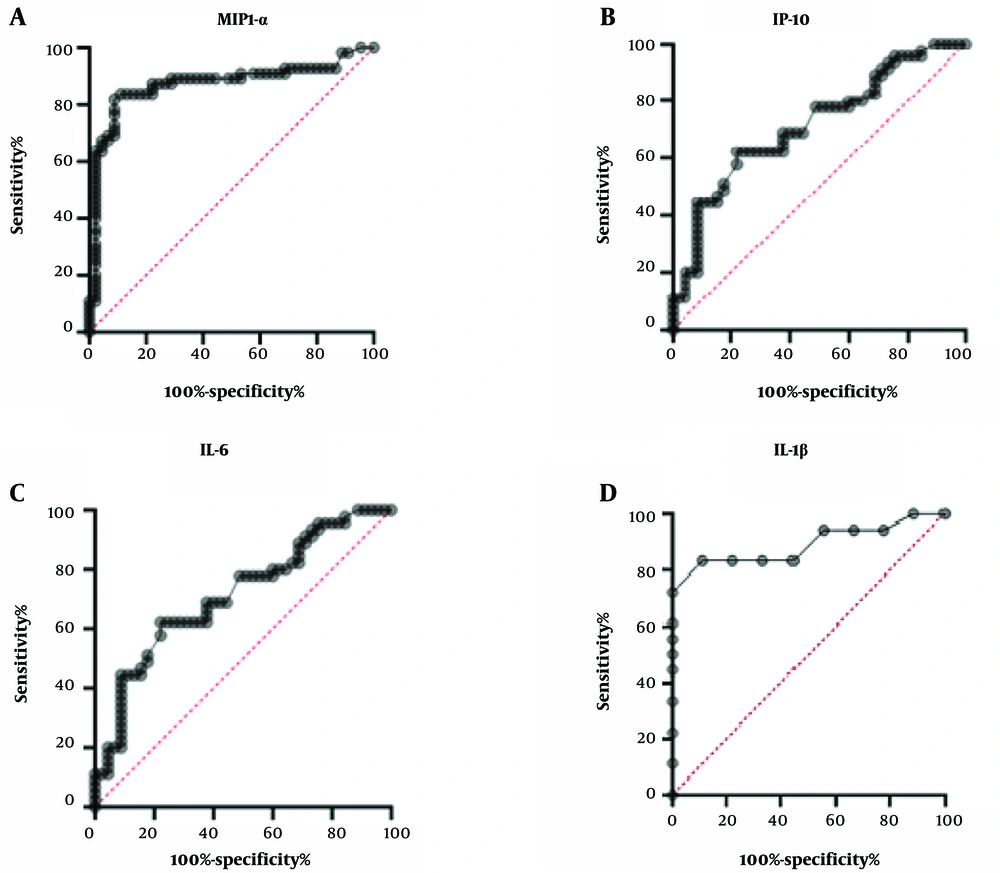
We then calculated the correlation to determine if there is any link between cytokines and chemokines. Interestingly, we found a strong positive linear association between MIP1-α and IP-10 in patients with severe infection (r = 0.86, P < 0.05). Notably, no correlations were observed between PD-1 and MCP-1 in mild and recovered COVID-19-infected patients. The diagnostic value of cytokines and chemokines for illness severity was assessed using the receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) (Figure 3). The AUC values of ROC for MIP1-α, IP-10, IL-6, and IL-1β were 0.79, 0.71, 0.93, and 0.89, respectively. These results indicate that these cytokines could predict the disease severity of COVID-19 infections (Figure 3). Our data suggest that these cytokines and chemokines may serve as biomarkers for disease severity in COVID-19-infected patients.
5. Discussion
The severity of COVID-19 has been linked to increased inflammation caused by a significant release of proinflammatory elements (9, 11). The rise in inflammation indicators is the key factor underlying the systemic vasculitic processes and abnormalities in coagulation processes that result in most parenchymal lesions in vital organs (12). We have identified that elevated levels of MIP1-α, IP-10, IL-6, and IL-1β during SARS-CoV-2 acute infection are associated with severe COVID-19. Cytokine levels can exhibit significant variations throughout the course of COVID-19, depending on factors such as the disease phase, the use of immunomodulatory medications, or the inherent characteristics of patients (13). Elevated levels of IL-6 have consistently been associated with severe cases of COVID-19 (14-16), playing a crucial role in the cytokine storm triggered by SARS-CoV-2 infection, which contributes to organ failure and severe lung damage (17-19).
In line with these findings, our study discovered a potential link between IL-6 and the severity of COVID-19. According to Ponti et al., 2020, there was a notable increase in IL-6 levels among non-survivors compared to survivors, as well as among individuals with severe disease compared to those with non-severe disease (12). Notably, IL-6 serves important biological functions, including the promotion of B cell differentiation into IgM and IgG and the stimulation of Th17 cell development and function (20). Multiple studies provide compelling evidence that IL-6 levels can serve as a valuable predictive factor for assessing the risk of severe disease and mortality (21, 22). Furthermore, large observational studies have demonstrated the beneficial effects of IL-6 inhibitors (23-25).
IL-1β, a potent proinflammatory cytokine essential for host defense responses against infections, can also exacerbate damage in chronic diseases and acute tissue injuries. Numerous studies have established a correlation between IL-1β and COVID-19, with some suggesting that blocking its release could potentially improve patients' condition (26). Consistent with these findings, our study also identified a link between IL-1β and the severity of COVID-19. Interestingly, the elevated levels of IL-1β observed at month 1 in COVID-19 patients may not necessarily be attributed solely to increased reactive oxygen species (ROS) levels (27). Viral antigens could initiate the inflammasome pathway, contributing to the sustained production of IL-1β (28). In vitro experiments conducted by Bertoni et al. demonstrated the activation of the NLRP3 inflammasome by the SARS-CoV-2 protein ORF3a (29). Furthermore, it has been shown that the SARS-CoV-2 nucleocapsid protein (N) can directly interact with NLRP3, promoting inflammasome assembly and activation, resulting in the production of various inflammatory molecules in mice (30).
Overall, these findings highlight the significance of IL-1β in the context of COVID-19 and shed light on potential mechanisms underlying its involvement in disease progression. Chemokines play a pivotal role in regulating leukocyte trafficking by stimulating adaptive immunity, recruiting and activating lymphocytes at the site of infection, and modulating the T-helper type-1 (Th1) or Th2 response (31). In our study, we observed significantly higher levels of MIP-1α in patients with severe COVID-19 compared to those with mild symptoms and healthy individuals. MIP-1α has been extensively studied for its significant immuno-adjuvant activity, acting as a chemoattractant for inflammatory cells such as immature DCs, macrophages, and monocytes (32). Moreover, MIP-1α stimulation enhances the production of IFN-γ, which is crucial for acquiring Th1 immunity. Animal models have demonstrated the vital role of MIP-1α in mediating virus-induced inflammation, as established several decades ago (33, 34).
Notably, MIP-1α, along with MIP-3β, holds great importance in modulating the efficacy and polarization of antigen-specific immunity (35). Additionally, studies have shown that IL-1β can enhance the production of MIP-1α through the activation of NF-κB (27). These findings collectively underscore the significance of MIP-1α in COVID-19 and its association with immune responses and inflammation mediated by other cytokines. Furthermore, our study revealed a significant association between serum levels of IP-10 and disease severity, suggesting its potential as a biomarker for predicting disease progression (5, 36, 37). These findings align with previous studies that have reported a marked elevation of IP-10 in both the blood and lung tissue of SARS patients (38). IP-10 plays a crucial role as a mediator of monocyte/macrophage-induced T cell activation and has been implicated in the pathogenesis of COVID-19 (39-41). IP-10 has been found to have pro-inflammatory and anti-angiogenic properties. It has been suggested as a potential connector between inflammation and angiogenesis in COVID-19 patients (42, 43). Interestingly, a corresponding increase in both IP-10 and MIP-1α was observed in patients who were transferred to the intensive care unit (ICU) during hospitalization, indicating a worsening clinical condition (44).
While plasma inflammatory cytokines exhibit high heterogeneity and serve a wide range of biological functions, they hold significant value as biomarkers for the diagnosis, management, and prognosis of various inflammatory diseases (45). It is important to acknowledge that different studies exploring cytokine assessment as predictors of severity have yielded varying results. These differences could be attributed to factors such as the use of different quantification methods, kits, relatively small sample sizes in some studies, and variations in the populations under investigation. Therefore, further studies are warranted to validate these biomarkers. In conclusion, our study aimed to compare the cytokine/chemokine profiles of severe and mild COVID-19 patients with those of healthy individuals. The results revealed that MIP1-α, IP-10, IL-6, and IL-1β exhibited significant predictive potential for the disease and its severity.
5.1. Conclusions
These findings provide valuable insights into the underlying mechanisms of COVID-19 progression and offer potential avenues for novel therapeutic approaches to address the cytokine storm observed in COVID-19. These approaches could involve globally targeting inflammation or neutralizing specific key inflammatory mediators. It is suggested to conduct studies with other markers and with larger sample sizes to obtain more comprehensive results in the future.