1. Background
Human immunodeficiency virus type 1 (HIV-1) remains a major global public health concern, affecting millions worldwide. As of the end of 2022, the World Health Organization (WHO) estimated that approximately 39 million people were living with HIV-1, with AIDS-related illnesses resulting in nearly 630,000 deaths (1). Despite significant advancements in antiretroviral therapy (ART) over the past decade, which have markedly improved the well-being and longevity of HIV-infected patients (2), the development of drug-resistant HIV-1 variants continues to pose challenges to effective treatment (3). A multitude of antiretroviral drugs have been developed through intensive efforts by experts in fields such as retrovirology, medicinal chemistry, enzymology, computational modeling, and structural biology (4). These drugs target several stages of the viral lifecycle, including cell fusion and entry, as well as the activities of the three viral enzymes: Protease (PR), reverse transcriptase (RT), and integrase (IN) (5).
The HIV-1 PR is crucial for the proteolytic processing of the viral polyprotein, leading to the cleavage of peptides that fold into functional proteins essential for the viral lifecycle (6). In combination therapy, these antiviral agents have proven highly effective. However, the virus can evolve, exhibiting increasingly higher levels of resistance during drug treatment by combining major mutations with additional accessory or compensatory mutations that can persist even after the cessation of treatment (7). The emergence of antiviral drug resistance is influenced by several factors, such as low adherence rates, inappropriate ART regimens, incorrect medication prescriptions, toxicity, high pill burden, and genomic mutations (8). Genotypic resistance testing is crucial for identifying drug-resistant mutations in HIV-1 patients, providing valuable information for clinicians to consider before initiating or adjusting ART regimens (9).
2. Objectives
As the first study of its kind in this region, which has one of the highest rates of HIV-1 infection in the country, it is critical to identify factors that affect treatment outcomes for this population (10). Additionally, we aim to identify the most common drug-resistant mutations and assess the level of susceptibility of different PIs in this area. Notably, the prevalence of HIV-1 drug resistance in Iran is reported to range from 5% to 15% (11-13). Our findings are expected to have significant implications for the management and treatment of HIV-1 patients and contribute to efforts to control and prevent the spread of drug-resistant HIV-1 strains.
3. Methods
3.1. Study Design
In this cross-sectional study, 59 HIV-1-positive patient samples were randomly obtained from health centers in Lorestan province, without prior knowledge of drug-resistance patterns. Samples were collected between 2015 and 2016. To address the main goal of the study, patient selection was based on two characteristics: Those receiving ART and those who were drug-naive. Twenty-seven patients were placed in the drug-naive group, naive for all drug classes, and 32 patients were in the group that had received ART. Demographic and clinical information such as age, sex, CD4 count, and viral load measurements are presented in Table 1. We included samples from patients aged 16 years and older for both groups. Patients who had been on antiretroviral regimens for at least one year were included, with treatment information provided by the physicians.
| Characteristics of Patients | Naive Patients | Antiretroviral Experienced Patients | Total |
|---|---|---|---|
| Sex | |||
| Male | 16 (47) | 18 (53) | 34 (100) |
| Female | 11 (44) | 14 (56) | 25 (100) |
| Age groups (y) | |||
| 16 - 26 | 5 (45) | 6 (55) | 11 (100) |
| 27 - 36 | 11 (57.1) | 10 (42.9) | 21 (100) |
| 37 - 47 | 8 (42) | 11 (58) | 19 (100) |
| 48 - 57 | 3 (37.5) | 5 (62.5) | 8 (100) |
| Transmission route | |||
| Heterosexual contact | 8 (50) | 8 (50) | 16 (100) |
| Injecting drugs | 12 (44.4) | 15 (55.5) | 27 (100) |
| Prisoners with high-risk behaviors | 5 (45.4) | 6 (54.5) | 11 (100) |
| Unknown | 2 (40) | 3 (60) | 5 (100) |
| Laboratory parameters CD4 (cells/µL) | |||
| < 200 | 6 (40) | 9 (60) | 15 (100) |
| 200 - 349 | 12 (46) | 14 (54) | 26 (100) |
| 350 - 499 | 8 (57) | 6 (43) | 14 (100) |
| ≥ 500 | 1 (25) | 3 (75) | 4 (100) |
| HIV-1 viral load | |||
| RNA (copy/mL) | 17800 (1553 - 31200) | 66617 (13074 - 598568) | 27002 (1553 - 598568) |
a Values are expressed as No. (%) or [median (min - max) × 106].
3.2. RNA Extraction, Amplification, and Genotyping
The first step involved extracting viral RNAs from plasma following the manufacturer’s instructions (QIAamp Viral RNA Mini Kit, Qiagen). Then, cDNA was synthesized using a one-step kit (Invitrogen) at 50°C for 30 minutes as recommended by the manufacturer. The HIV-1 PR gene fragments were amplified using nested-PCR with specific primers. The first round of PCR was conducted in a total volume of 25 µL, including 5 µL of cDNA, 12.5 µL of master mix 2x (Amplicon), 1 µL of each PCR primer (10 pM), and 5.5 µL of DEPC-treated water. The final amplified region covered the HIV PR gene from position 1800 to 2125 of the reference gene (accession number: NC_001802).
The following primer pairs were used (14): Outer forward primer (5’-GAGCCAACAGCCCCACCAG-3’); outer reverse primer (5’-GCCATTGTTTAACYTTTGGGCCATCC-3’); inner forward primer (5’-CTCARATCACTCTTTGGCAACG-3’); inner reverse primer (5’-CTGGTAYAGTHTCAATAGGRCTAAT-3’). The cycle steps included annealing at 55ºC, extension at 72ºC, and denaturation at 94ºC, each for 1 minute. The outer PCR consisted of 45 cycles, and the nested PCR had 40 cycles. A 5-minute final extension step at 72ºC was performed. Final PCR products (325 bp) were observed using 2% agarose gel electrophoresis, and positive samples were sequenced by Sanger sequencing using 3130 genetic analyzers (Applied Biosystems).
3.3. Analysis of Drug Resistance and Phylogenetic Relationships from Protease Sequence
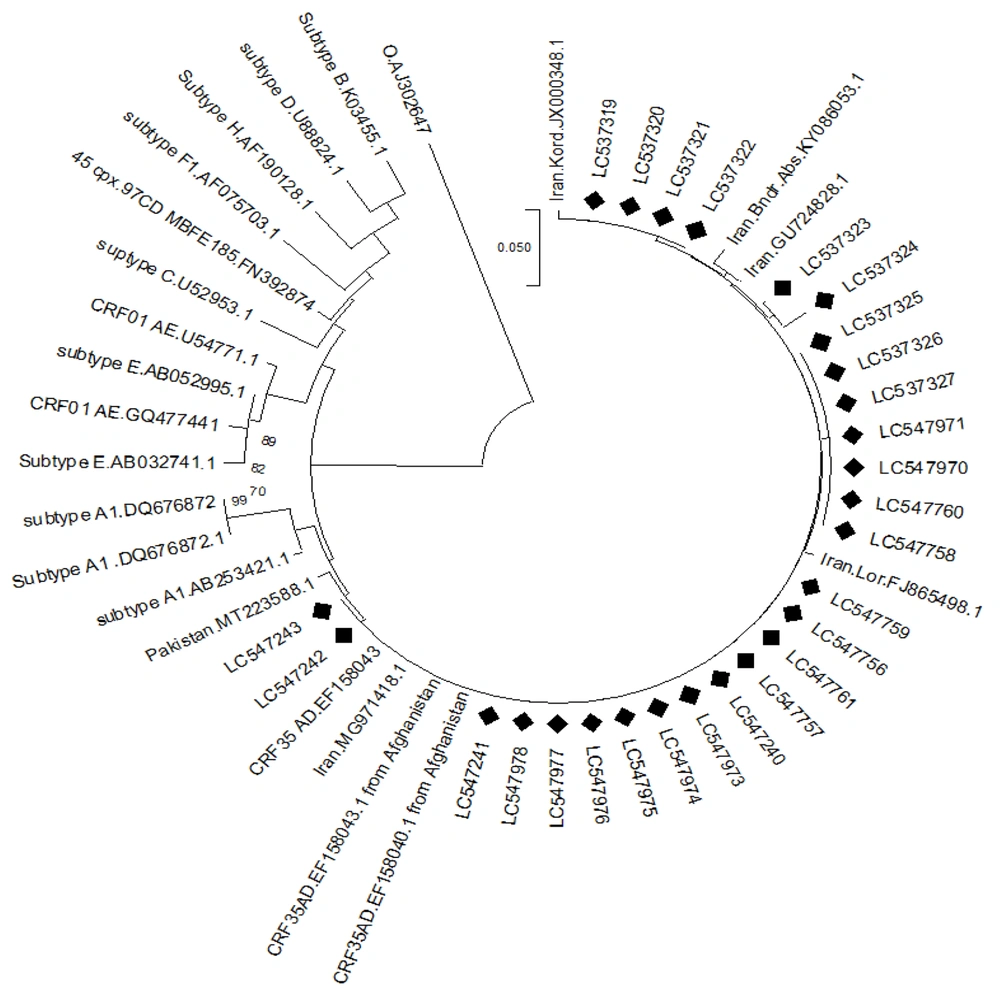
BioEdit software (version 7.2.5) was used to correct the sequences. The Stanford University HIV Drug Resistance mutations database [version 4.2.6 (http://hivdb.stanford.edu)] was utilized to evaluate the PR sequences for drug resistance in patients. The phylogenetic tree was constructed using molecular evolutionary genetics analysis (MEGA) software, version 11.0.13, employing the neighbor-joining method, 1000 bootstrap replicates, and the Tamura-Nei model. The PR nucleotide sequences from this study were submitted to the GenBank database, with the relevant accession numbers as shown in Figure 1.
3.4. Statistical Analysis
Data analysis was conducted using SPSS version 20 software (SPSS Inc., Chicago, IL). The normality of quantitative variables was assessed with the Kolmogorov-Smirnov test. Continuous variables were analyzed using the Kruskal-Wallis test. Statistical differences between the two groups were evaluated using the Fisher exact test and the chi-squared test. A P-value of less than 0.05 was deemed statistically significant.
4. Results
4.1. Characteristics of Participants
Fifty-nine HIV-positive patients were enrolled in the study: Twenty seven were drug-naive, having not taken any ART regimen, and 32 had been on ART for at least 12 months. Of the participants, 34 (57.6%) were men and 25 (42.4%) were women (Table 1). The mean age of all patients was 38.4 years [standard deviation (SD) = 11.6]. Participants were classified into four age groups: 16 – 26 (18.6%), 27 – 36 (35.6%), 37 – 47 (32.3%), and 48 – 57 years old (13.5%). Human immunodeficiency virus transmission primarily occurred through injecting drug use (45.7%), followed by heterosexual contact (27.1%), and prisoners with high-risk behaviors (18.6%). Although all participants had been on ART for at least one year, 9 (28.1%) of them had a CD4 count of less than 200 cells/mm³. The mean CD4 count was 341 cells/mm³ for drug-naive patients and 278 cells/mm³ for drug-experienced patients. The baseline characteristics of the study groups are shown in Table 1.
4.2. Monitoring the Prevalence of Drug-Resistant Mutations in Treatment-Naive HIV-1-Infected Individuals
In this study, which comprised 59 participants, our focus was narrowed down to 27 individuals who underwent sharp band amplification in PCR, followed by sequencing and drug-resistance analysis. It was found that a significant majority of 96.2% (26 out of 27) of HIV-1 positive participants carried at least one drug-resistant mutation. Additionally, 6 patients (22%) had multiple mutations. Of the 27 samples analyzed, 14 cases were from the ART-experienced group, while the remaining 13 were from the drug-naive group. All patients on ART exhibited mutations that suggested drug resistance related to PIs (Table 2).
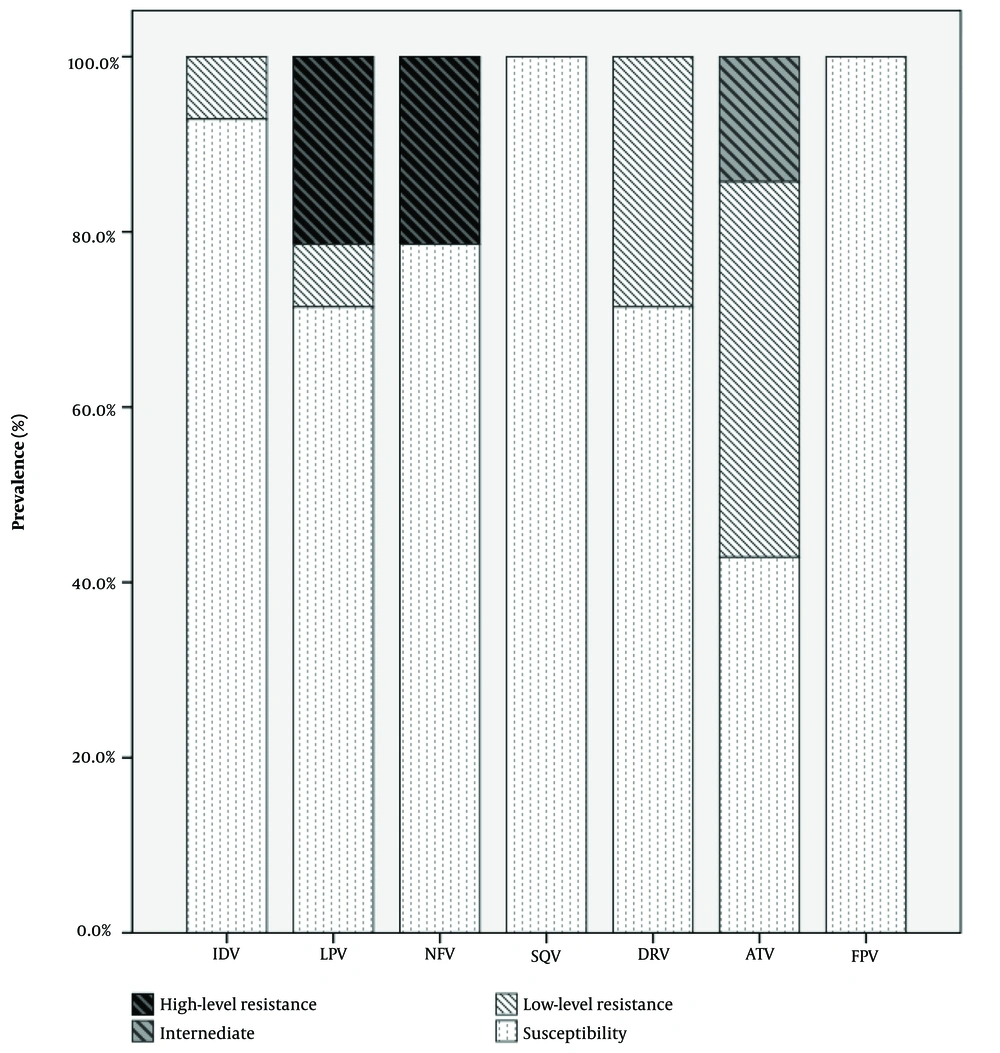
Among the 14 patients who had taken antiretroviral drugs, 11 (78%) had major mutations, while 3 (22%) had minor mutations. The most common major PI mutations in the antiretroviral-experienced patients were D30N (27.2%, 3.11) and V32I (27.2%, 3.11), followed by G48A (18.1%, 2.11), L90M (18.1%, 2.11), and L76V (9%, 1.11). The most frequent minor PI mutations in this group were K20R (40%, 2.5), L10I (20%, 1.5), F53I (20%, 1.5), and V11I (20%, 1.5). No major PI mutations were found in drug - naive patients. K20R (85.7%) and L10I (14.2%) were observed as minor PI mutations in drug-naive patients. We did not find significant drug resistance in the naive patient group (Table 2). Table 3 lists the PI agents and their levels of susceptibility in drug-experienced patients. The highest percentages of high-level resistance were observed with lopinavir (LPV) and nelfinavir (NFV), both at 50%. Intermediate-level resistance was noted for atazanavir (ATV), which also showed the highest percentage (50%) in the low-level resistance category. As shown in Figure 2, the susceptibility of patients to saquinavir (SQV) and fosamprenavir (FPV) was higher than others.
| Patients Taking Antiretroviral Therapy | Naive Patients | |
|---|---|---|
| Minor | Major | Minor |
| K20R, 2 (40) | D30N, 3 (27.2) | K20R, 12 ( 85.7) |
| F53l, 1 (20) | G48A, 2 (18.1) | L10I, 2 (14.2) |
| L10I, 1 (20) | L76V, 1 (9) | |
| V11I, 1 (20) | L90M, 2 (18.1) | |
| V32I, 3 (27.2) | ||
| Total: 5 (100) | 11 (100) | 14 (100) |
a Values are expressed as No. (%).
| Medication | Patients Taking Antiretroviral Therapy | Susceptibility | ||
|---|---|---|---|---|
| High | Intermediate | Low | ||
| IDV | 0 (0) | 0 (0) | 1 (8) | 13 (92.8) |
| LPV | 3 (50) | 0 (0) | 1 (8) | 10 (71.4) |
| NFV | 3 (50) | 0 (0) | 0 (0) | 11 (78.5) |
| SQV | 0 (0) | 0 (0) | 0 (0) | 14 (100) |
| DRV | 0 (0) | 0 (0) | 4 (33) | 10 (71.4) |
| ATV | 0 (0) | 2 (100) | 6 (50) | 6 (42.8) |
| FPV | 0 (0) | 0 (0) | 0 (0) | 14 (100) |
Abbreviations: LPV, lopinavir; SQV, saquinavir; IDV, indinavir; NFV, nelfinavir; ATV, atazanavir; DRV, darunavir; FPV, fosamprenavir.
aValues are expressed as No. (%).
4.3. Human Immunodeficiency Virus Subtyping
The phylogenetic tree of the PR region is shown in Figure 1. These analyses indicated that all 27 participants (100%) were infected with CRF35_AD, which is the dominant HIV-1 subtype in Iran.
5. Discussion
Drug resistance is a significant issue in HIV-positive patients that can greatly affect treatment outcomes. The development of resistance can arise from a range of factors, including inadequate adherence to ART regimens and the emergence of resistant strains of HIV during treatment. Protease inhibitors are particularly prone to resistance, underscoring the importance of understanding and monitoring their effectiveness (15). Nine PR drugs have been approved for clinical AIDS treatment, including SQV, indinavir (IDV), ritonavir (RTV), NFV, amprenavir (APV), LPV, ATV, tipranavir (TPV), and darunavir (DRV). While some studies suggest that resistance to PIs is not as common as resistance to other antiretroviral drugs, others have found that resistance can emerge within weeks to months of starting treatment, with PI resistance becoming increasingly prevalent in HIV-positive patients (16, 17). For instance, a recent study by Obsa et al. identified that out of 56 patients analyzed, 14 (25%) had PI resistance mutations, significantly higher than figures recorded for non-nucleoside RT inhibitors (NNRTIs) (5/56, 9%) and IN inhibitors (2/56, 4%) (18).
Resistance to PIs can be attributed to the rapid mutation rate of HIV-1 PR, producing approximately 108 virions per day with an error probability of 5/10,000 bases. This rapid mutation leads to the development of drug-resistant mutants that form weakened bonds with PIs. Moreover, numerous studies have shown that PR mutations accumulate over time, causing the virus to become less responsive to inhibitors (19, 20). Host genetic factors, such as human leukocyte antigen (HLA) alleles and other immune-related genes like CCR5 and interferon-induced genes, also play a crucial role in resistance development (21, 22). The results of a cohort study conducted on HIV patients undergoing ART suggest that PIs should be prescribed as first-line drugs in ART regimens for Iranian patients who experience virological failure (23). Additionally, a study carried out in Iran reported that the prevalence of PI mutations was 9.1%, with only three minor mutations (L10I, L10V, and G73S) found (24). Understanding why and how resistance develops is particularly important in developing new therapeutic strategies for HIV.
Our study revealed that the majority of HIV-1-positive patients harbored mutations in the PR gene. Specifically, 26 (96.2%) of the participants had at least one drug-resistant mutation, and 6 (22%) had more than one mutation. Among 14 patients with ART experience, 11 (78%) had major mutations, whereas 3 (22%) had minor mutations. These findings are consistent with previous studies demonstrating the high prevalence of drug resistance mutations among HIV-1-positive patients in Iran (25, 26). For instance, Sadeghi et al. conducted a study where they found that out of 17 patients, 2 (12%) had major PI resistance mutations, while 7 (41%) had minor PI resistance mutations (27). Another study conducted in Iran showed that 32% of patients undergoing antiretroviral treatment had mutations related to PI drug resistance (28). However, our study revealed a higher prevalence of the PR gene compared to other reports. This finding suggests that the region studied may have a larger population of HIV-positive patients, leading to increased virus circulation throughout society. With an increased replication rate, the virus mutates more frequently, potentially leading to the development of drug-resistant strains.
Numerous PR mutants have been thoroughly characterized, encompassing approximately 20 distinct mutations that result in significantly reduced susceptibility to clinical PIs. The majority of significant resistance-associated mutations (RAMs) related to PIs are found within various structural components of the active site pocket. These include the active site loop (comprising residues D30, V32, and L33), the 80s loop (encompassing residues V82 and I84), which together delineate the pocket’s boundaries, and the flap region of the PR (involving residues M46, I47, G48, I50, and I54) (29). Over half of the PR residues are implicated in resistance to all currently available clinical PIs, leading to diminished efficacy and potential treatment failure (6). The emergence of major mutations engenders drug resistance through three primary molecular mechanisms: Alterations of residues within the inhibitor binding site can directly impact PR–PI interactions; certain distal mutations induce resistance by modifying dimerization through changes in intersubunit contacts; and other distal mutations, such as L76V, can destabilize the dimer, thereby reducing PI effectiveness due to structural rearrangements at crucial PR sites (29).
This study also demonstrated that the most prevalent major PI mutation in patients with prior antiretroviral exposure was D30N, followed by V32I, G48A, L90M, and L76V. Conversely, no major PI mutations were detected in drug-naive individuals. Additionally, the study highlighted that the most common minor PI mutations in patients with antiretroviral experience were K20R, L10I, F53L, and V11I. When devising the optimal therapeutic strategy for a patient, resistance constitutes one of several critical considerations. The D30N mutation, a primary alteration associated exclusively with resistance to NFV, highlights this inhibitor’s specificity (30). In contrast, V32I is linked to increased resistance to LPV. L90M, a primary mutation, is implicated in resistance to multiple PIs, including NFV, IDV, and SQV. The L76V mutation, when combined with frequent accompanying substitutions such as M46I, I54V, V82A, and L90M, significantly amplifies LPV resistance (31).
Notably, the G48A mutation, although exceedingly rare, may not induce polymorphic PI-selected mutations that augment resistance to DRV (32). The evolution of resistance to DRV necessitates 10 – 20 amino acid changes. Furthermore, it is plausible that distal mutations could influence dimerization or interactions with other HIV-1 or host proteins, potentially altering PR dynamics (33). Our study shows a high-level resistance to LPV and NFV, moderate-level resistance to ATV, while it is susceptible to SQV and FPV. These results are in line with recent studies on Iranian HIV-positive patients undergoing ART (34, 35).
Furthermore, the study revealed that all patients in Lorestan province were infected with the CRF35-AD strain, which is the dominant HIV-1 subtype in Iran. This is consistent with previous research that has also identified CRF35-AD as the predominant subtype in Iran (24, 25). The study's results raise concerns about the potential spread of drug-resistant strains of CRF35-AD in the region. It is imperative to acknowledge certain limitations within this study. The phylogenetic analysis conducted is restricted to the PR region of the viral genome. For a more holistic view of viral resistance patterns, it is crucial to also consider and sequence other significant regions, including gag and env. Furthermore, the absence of pediatric patient samples in our dataset may affect the breadth of our conclusions. Nevertheless, the data presented herein provide important insights into resistance mutations, which are valuable despite the methodological limitations. Future research with a larger cohort would be beneficial to reinforce the validity and robustness of these findings.
5.1. Conclusions
The development of drug resistance mutations in HIV is predominantly driven by the pressures of antiretroviral treatment. This dynamic leads to the formation of resistant viral variants that can be transmitted to treatment-naive individuals, resulting in expedited virological failure and a reduction in viable treatment options. It is imperative to accurately determine the prevalence, emergence, and transmission of drug resistance to effectively manage patient treatment and inform health policy decisions. In this study, we have documented a significant number of mutations in the PR gene among HIV-positive patients from Lorestan province, Iran, who have been treated with antiretrovirals. These findings underscore the critical need for meticulous selection of antiretroviral regimens (ARVs), ongoing patient follow-up, reinforcement of adherence strategies, and comprehensive monitoring of viral load to detect early signs of treatment failure.