1. Background
Shigellosis is a type of diarrhea known as bacillary dysentery caused by the Gram-negative rod-shaped bacterium Shigella spp. (1). This type of diarrhea can progress to a more severe form containing bloody mucus, which is associated with morbidity and mortality, especially in young children in developing countries (2). The infectious dose of Shigella spp. can be very low, as few as 10 organisms, facilitating person-to-person transmission (3). The Shigella genus consists of four subgroups based on biochemical and serological properties: (A) (Shigelladysenteriae), (B) (S.flexneri), (C) (S. boydii), and (D) (S. sonnei). Each subgroup can cause shigellosis, but S. flexneri and S. sonnei are the two major causes of the disease, especially in developing and industrialized countries (4). Antibiotics can help shorten the duration of illness and reduce the risk of spreading Shigella bacteria when treating patients with shigellosis. Several different types of antibiotics are effective in treating this infection (5).
Fluoroquinolones, cephalosporins, and sulfonamides are the drugs typically used to treat acute diarrhea caused by Shigella spp., according to current medical guidelines (6). However, the overuse of antibiotics, along with the emergence of Shigella strains that are resistant to multiple drugs, has reduced the number of treatment options for Shigella infections due to various resistance mechanisms (7-9). There are different mechanisms by which Shigella spp. can develop drug resistance, such as drug efflux, decreased cellular permeability, drug modification or inactivation by producing enzymes, and drug target mutations (10, 11). The main reason gram-negative bacteria are resistant to beta-lactam antibiotics is the production of extended-spectrum beta-lactamase enzymes (ESBLs). These enzymes can break down penicillins and first-, second-, and third-generation cephalosporins (12).
The distribution of virulence genes among different types of Shigella spp. can cause varying symptoms and outcomes (13). The Ipa and ial genes are important in helping Shigella cells invade the intestinal epithelium (14). The VirF and VirB (InvE) genes are involved in regulating invasion-related genes (15). SPATE genes are a group of genes that can be divided into class 1 and class 2. SPATE class 1 genes include sigA and sat, and SPATE class 2 genes include sepA and pic. These genes work together to allow Shigella to survive and cause colonization (16).
Karimi Yazdi et al., in their study focused on pediatric patients under 14 years of age in Iran, found that S. sonnei was the dominant species isolated from children with shigellosis, confirming the findings of Abbasi et al.. These kinds of studies highlight the high prevalence of multidrug resistance among Shigella isolates in Iran and emphasize the importance of continuous monitoring of antibiotic resistance patterns in different regions of the country (17, 18).
2. Objectives
Understanding the antibiotic resistance pattern and the relationship between virulence genes and antimicrobial resistance in Shigella spp. is critical for developing new and effective treatments for shigellosis. In this study, after determining the antibiotic resistance pattern of Shigella clinical isolates to different antibiotic groups, the presence of antibiotic resistance genes (ARGs), including blaCTX-M1, blaCTX-M2, blaCTX-M8, blaSHV, blaTEM, and blaOXA in Shigella spp., was investigated. Finally, the distribution of five virulence genes (ial, virF, invE, sigA, and pic) in the studied Shigella isolates was evaluated.
3. Methods
3.1. Isolation and Identification of Shigella spp.
In this cross-sectional study, 60 Shigella isolates were collected from the microbiology laboratories of two hospitals (Imam Khomeini and Milad) in Tehran, Iran, from February 2023 to June 2023. These isolates had been previously obtained from stool samples of patients ranging in age from under one year to 32 years old. Shigella isolates were checked using standard biochemical tests (urease, carbohydrate fermentation test, H2S, methyl red, motility, citrate utilization, and indole) and molecular tests (amplifying the ipaH gene) (6). All isolates were then maintained in Brain Heart Infusion (BHI) Broth containing 20% glycerol at -70°C.
3.2. Antimicrobial Susceptibility Testing
The antibiotic susceptibility of Shigella isolates was assessed using the Kirby-Bauer disk diffusion technique on Muller Hinton agar (MHA; Merck, Darmstadt, Germany) according to the Clinical and Laboratory Standards Institute (CLSI, 2021) guidelines (19). Ten different antibiotic disks (Padtanteb, Iran), representing eight classes, were used, including aminoglycosides (amikacin-30 μg, kanamycin-30 μg, and gentamicin-10 μg), carbapenems (imipenem-10 μg), macrolides (azithromycin-15 μg), tetracyclines (tetracycline-30 μg), penicillins (ampicillin-10 μg and amoxicillin-25 μg), fluoroquinolones (nalidixic acid-30 μg and norfloxacin-10 μg), cephalosporins (cefoxitin-5 μg and cefepime-30 μg), and chloramphenicol-30 μg. Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 strains were used as reference strains. Isolates resistant to more than three classes of antibiotics were classified as multidrug-resistant (MDR) (20), and the following formula (21) was used to determine the multiple antibiotic resistance (MAR) index of isolates: MAR = (The number of antimicrobial agents to which a particular Shigella isolate shows resistance/the total number of antibiotics that were utilized in treating an isolate).
3.3. Molecular Detection of Beta-lactamase Resistance Genes and Virulence Genes Using Multiplex PCR
3.3.1. DNA Extraction
Genomic DNA was extracted from the bacterial samples using a commercial Genomic DNA extraction Kit (cat No: DM05050, Gene Transfer Pioneer, Pishgaman Co, Iran). The extraction procedure was performed according to the manufacturer's instructions. DNA concentration and quality were determined with a NanoDrop 2000 Spectrophotometer (Thermo Fisher, USA) and examined by gel electrophoresis.
3.3.2. Multiplex PCR Technique
The beta-lactamase genes blaCTX-M1, blaCTX-M2, blaCTX-M8, blaSHV, blaTEM, and blaOXA were simultaneously investigated using specific primers and two sets of multiplex PCR techniques (Table 1). Furthermore, the ipaH gene encoding an E3 ubiquitin-ligase in the Shigella family was considered an internal positive control (Table 1). The validity of all primers was checked using the Blast databases available from NCBI, and they were synthesized by the oligonucleotide DNA synthesis service of Macrogen Inc., South Korea. Positive controls were sequencing confirmed and applied for the targeted beta-lactamase genes. A PCR reaction mixture was prepared by blending 2 μL of DNA template (concentration 50 ng/μL) with 10 μL of 2X master mix containing the standard buffer and 0.7 μL of each of the six primer pairs. This PCR mixture had a final volume of 20 μL.
| Genes Group | Primer Sequence (5' - 3') | PCR Product Length (bp) | Reference |
|---|---|---|---|
| β-lactamase | |||
| TEM | F: TCGGGAAATGTGCG | 857 | (22) |
| R: GGGTTTGATACCGGCACCCGT | |||
| OXA | F: GCAGCGCCAGTGCATCAAC | 198 | (23) |
| R: CCGCATCAAATGCCATAAGTG | |||
| SHV | F: GCCGGGTTATTCTTATTTGTCGC | 768 | (24) |
| R: ATGCCGCCGCCAGTCA | |||
| CTX-M1 | F: CGTCACGCTGTTGTTAGGAA | 593 | (25) |
| R: ACGGCTTTCTGCCTTAGGTT | |||
| CTX-M2 | F: TTAATGATGATCTCAGAGCATTC | 901 | (26) |
| R: GATACCTCGCTCCATTTATTG | |||
| CTX-M8 | F: CGCTTTGCCATGTGCAGCACC | 307 | (26) |
| R: GCTCAGTACGATCGAGCC | |||
| Virulencefactors | |||
| Ial | F: GCTATAGCAGTGACATGG | 320 | (27) |
| R: ACGAGTTCGAAGCACTC | |||
| Vir | F: AGCTCAGGCAATGAAACTTTGAC | 618 | (27) |
| R: TGGGCTTGATATTCCGATAAGTC | |||
| InvE | F: CGATAGATGGCGAGAAATTATATCCCG | 766 | (28) |
| R: CGATCAAGAATCCCTAACAGAAGAATCA | |||
| SigA | F: CCGACTTCTCACTTTCTCCCG | 430 | (27) |
| R: CCATCCAGCTGCATAGTGTTTG | |||
| Pic | F: ACTGGATCTTAAGGCTCAGGAT | 572 | (27) |
| R: GACTTAATGTCACTGTTCAGCG | |||
| Ipa | |||
| IpaH | F: GCTGGAAAAACTCAGTGCCT | 423 | (29) |
| R: CCAGTCCGTAAATTCATTCT |
Thermal cycling conditions for PCR were as follows: Initial denaturation at 95°C for 3 min, followed by 30 cycles of denaturation at 95°C for 30 s. For the set of blaSHV, blaTEM, and blaOXA genes, the annealing temperature was set at 55.5°C for 30 seconds. For the set of blaCTX-M1,blaCTX-M2, and blaCTX-M8 genes, the annealing temperature was set at 57°C for 30 seconds. This was followed by a primer extension step at 72°C for 1 minute. After the last PCR cycle, an additional incubation step was included to complete the chain extension. This step included incubation at 72°C for 5 min. To analyze the PCR products, the multiplex PCR mixture was subjected to electrophoresis using 1.5% (w/v) agarose gel with a voltage of 110 V for 60 min. After the electrophoresis process, the DNA fragments in the gel were observed using a UV transilluminator (Protein Simple, Red imagen SA-1000). Two strains of S.sonnei ATCC 25931 and Escherichia coli ATCC 25922 were used as negative and positive controls, respectively.
To determine the distribution of virulence genes, including ial, virF, invE, sigA, and pic, a multiplex PCR assay and specific primers (Table 1) were applied. For this purpose, a new mixture was prepared. This 20 μL multiplex PCR mixture consisted of 2 μL of DNA template (concentration of 50 ng/μL), 10 μL of 2X master mix with the standard buffer, and 1 μL of each of the four primer pairs. The thermal cycling conditions were set as follows: Initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s. The next steps included primer annealing and extension. For the annealing step, the temperature was set at 58°C for 30 s. This was followed by a primer extension step at 72°C for 1 min. After the cycling conditions mentioned earlier, an additional incubation step was included to complete the chain extension. This step consisted of incubation at 72°C for 7 min following the last PCR cycle. Then, multiplex PCR products of three genes including virF, invE, and sigA were purified for sequencing and sent to Niagenenoor Corporation, Tehran, Iran, along with the used primers. Online BLAST software was employed to examine the sequences in the NCBI database (https://www.ncbi.nlm.nih.gov/BLAST/).
3.4. Statistical Analysis
SPSS statistical software version 22 was used for data analysis. The chi-square test and Cramer's V test were employed to determine the significance and strength of the associations between antibiotic resistance genes and the antibiotic resistance observed in the studied bacterial samples. The P-value was set at less than 0.05.
4. Results
4.1. Isolation and Identification of Shigella spp.
Sixty bacterial isolates were confirmed as Shigella spp. after performing the morphological, biochemical and molecular tests.
4.2. Antibiotic Resistance Pattern
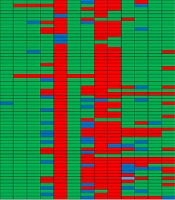
All Shigella isolates in this research were assessed separately for susceptibility to common clinically relevant antibiotics (Table 2 and Figure 1). Out of 60 isolates, 93% showed resistance to amoxicillin (56/60), 90% were resistant to tetracycline (54/60), and 80% were resistant to ampicillin (48/60). Resistance to nalidixic acid and chloramphenicol was observed in 33.3% (20/60) and 23.3% (14/60) of isolates, respectively. Twenty percent of isolates showed resistance to imipenem, cefepime, and azithromycin (12/60). Regarding other tested antibiotics, 11.7% were resistant to amikacin (7/60), 3.3% were resistant to kanamycin and gentamicin (2/60), and 1.7% showed resistance to norfloxacin and cefoxitin (1/60). Additionally, among the sixty isolates, 36.66% were determined to be MDR. The most common MDR pattern was simultaneous resistance to chloramphenicol, ampicillin, tetracycline, and amoxicillin (Table 3).
| Antibiotics | Shigella Isolates Total (n = 60) a | No. of Isolates | Valid Percent |
|---|---|---|---|
| Norfloxacin | 1.7 | 1 | 1.7 |
| Gentamicin | 3.3 | 2 | 3.3 |
| Kanamycin | 3.3 | 2 | 3.3 |
| Cefepime | 20 | 12 | 20 |
| Amoxicillin | 93.3 | 56 | 93.3 |
| Cefoxitin | 1.7 | 1 | 1.7 |
| Nalidixic acid | 33.3 | 20 | 33.3 |
| Tetracycline | 90 | 54 | 90 |
| Ampicillin | 80 | 48 | 80 |
| Azithromycin | 20 | 12 | 20 |
| Imipenem | 20 | 12 | 20 |
| Amikacin | 11.7 | 7 | 11.7 |
| Chloramphenicol | 23.3 | 14 | 23.3 |
a Values are expressed as (%).
| Pattern No. | Antibiotic Resistance Patterns | No. of Isolates | Overall MDR Isolates | Beta-lactamase Genes | Virulence Genes |
|---|---|---|---|---|---|
| 1 | C, AN, IPM, AZM, AM, TE, NA, AMX, FEP | 1 | 22/60 (36.66%) | blaCTX-M8, blaOXA | invE, virF, ial |
| 2 | AM, TE, NA, FOX, AMX, FEP, K, GM | 1 | blaCTX-M1, blaCTX-M8 | invE, virF, sigA | |
| 3 | C, AN, IPM, AZM, AM, TE, NA, AMX | 2 | -ND | invE, virF, sigA, ial | |
| 4 | C, IPM, TE, AM, NA, AMX, FEP | 1 | blaCTX-M1, blaCTX-M8 | virF, sigA | |
| 5 | C, AN, IPM, AM, TE, NA, AMX | 1 | - | invE, virF, sigA, pic | |
| 6 | C, AZM, AM, TE, NA, AMX | 2 | blaCTX-M1, blaCTX-M8 | invE, virF, sigA | |
| 7 | AN, IPM, AZM, AM, TE, AMX | 2 | blaCTX-M1 | invE, virF, sigA, pic | |
| 8 | AZM, TE, AM, NA, AMX | 1 | blaCTX-M1, blaCTX-M8 | virF, sigA | |
| 9 | AZM, TE, AM, AMX, FEP | 1 | blaCTX-M8 | virF, pic | |
| 10 | C, AM, TE, NA, AMX | 1 | blaCTX-M1, blaCTX-M8 | virF, sigA | |
| 11 | TE, NA, AMP, FEP | 1 | - | invE, virF, sigA, ial | |
| 12 | C, AM, TE, AMX | 4 | blaCTX-M1 | invE, virF, sigA | |
| 13 | C, AM, TE, NA | 1 | blaCTX-M1, blaCTX-M8 | virF | |
| 14 | AM, TE, NA, AMX | 1 | blaCTX-M1 | invE, virF, sigA, ial | |
| 15 | AM, TE, FEP, K | 1 | blaCTX-M1 | invE, virF, sigA | |
| 16 | AZM, NA, AMX | 1 | blaCTX-M8 | invE, virF, sigA |
Abbreviations: ND, no detected; NOR, norfloxacin; GM, gentamicin; K, kanamycin; FEP, cefepime; AMX, amoxicillin; FOX, cefoxitin; NA, nalidixic acid; TE, tetracycline; AM, ampicillin; AZM, azithromycin; IPM, imipenem; AN, amikacin; C, chloramphenicol.
a In this table, the antibiotic resistance patterns of 60 Shigella isolates are presented alongside their corresponding bla genes and virulence genes.
4.3. Antimicrobial Resistance Genes Identified in Shigella spp.
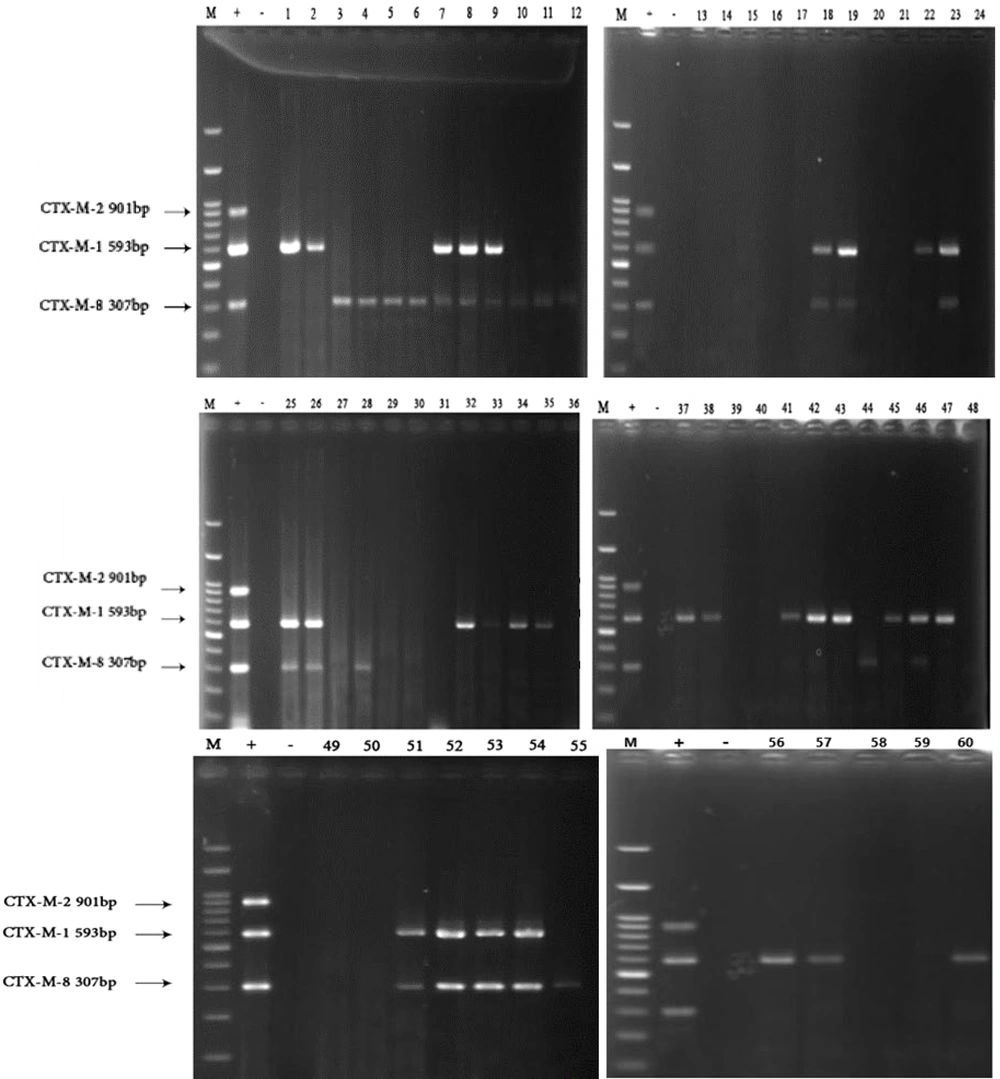
All 60 Shigella isolates were tested for the presence of the blaCTX-M1, blaCTX-M2, blaCTX-M8, blaSHV, blaTEM, and blaOXA genes. Figures 2 and 3 display the PCR gel electrophoresis results of these six resistance genes. All isolates were negative for blaCTX-M2 and blaSHV, but 30 (50%) isolates harbored blaCTX-M1. The frequency of the blaCTX-M8 gene was 38.3% (23 isolates). Only two (3.33%) isolates contained the blaTEM gene, and five isolates (8.33%) carried blaOXA. The blaCTX-M1 gene had a significantly higher frequency than the blaCTX-M8 gene. The results of multiplex PCR indicated the presence of 16 patterns in terms of carrying resistance genes in the 60 investigated Shigella isolates. Table 4 displays the occurrence of ARGs in Shigella isolates.
Multiplex PCR assay for detection of blaOXA, blaSHV, blaTEM, blaCTX-M1 and ipaH (as internal control) on 1.5% agarose gel. Lane 1, marker (100 bp); lane 2, positive control (Escherichia coli 25922 ATCC); lane 3, negative control (Shsonii 25931 ATCC); lane 1 – 60, extracted DNA of Shigella isolates.
| Antibiotic | Sh Isolates Total (n = 60) b | No. of Isolates | Valid Percent |
|---|---|---|---|
| blaTEM | 1.7 | 1 | 1.7 |
| blaSHV | 1.7 | 1 | 1.7 |
| blaOXA | 8.3 | 5 | 8.3 |
| blaCTX-M1 | 51.7 | 31 | 51.7 |
| blaCTX-M2 | 0.0 | 0 | 0.0 |
| blaCTX-M8 | 38.3 | 23 | 38.3 |
a In this table, the number of isolates and the percentage of isolates carrying each ARG are presented.
b Values are expressed as (%).
4.4. Distribution and Prevalence of Virulence Genes
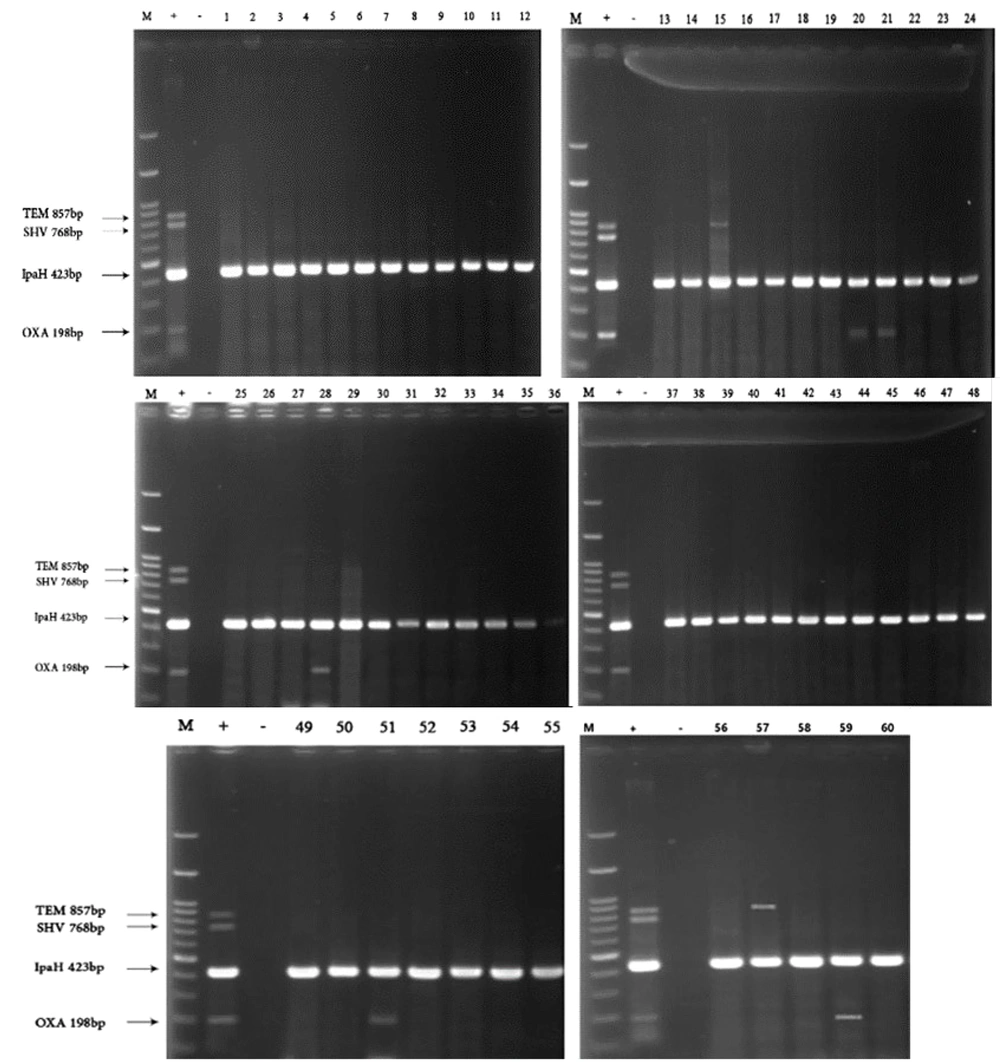
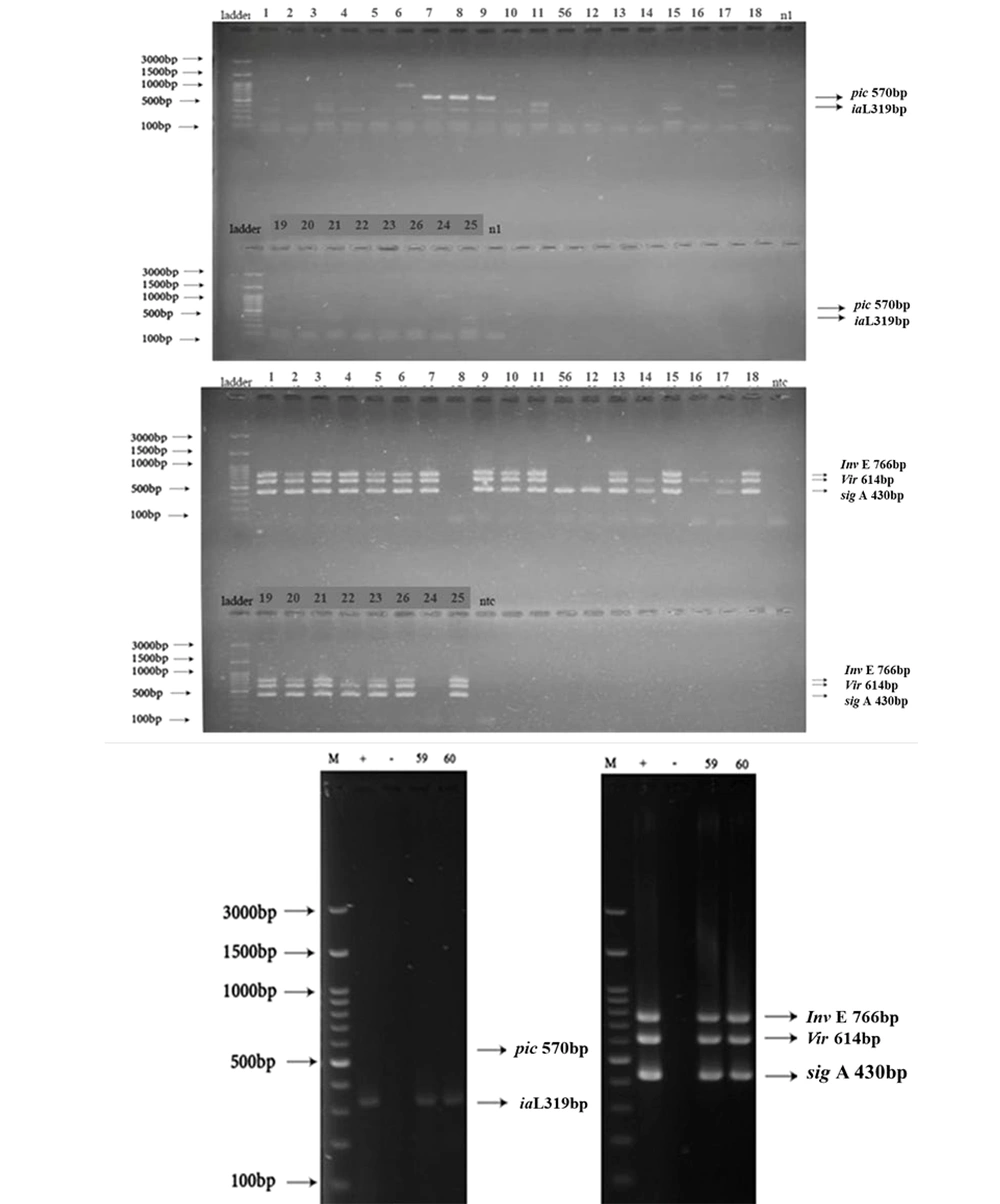
During the present investigation, all 60 Shigella isolates were subjected to PCR to detect five virulence genes, including ial, virF, invE, sigA, and pic. Figure 4 displays the PCR gel electrophoresis results of these five virulence genes. The distribution of virulence genes ial, virF, invE, sigA, and pic in the studied isolates was 28.3% (17/60), 85% (51/60), 68.3% (41/60), 81.7% (49/60), and 15% (9/60), respectively. Table 3 also shows the multidrug resistance profile of the Shigella isolates based on virulence genes.
Multiplex PCR assay for detection of A, pic (570 bp) and ial (319 bp); B, invE(766 bp),virF (614 bp) and sigA(430 bp); and C, pic (570 bp), ial (319 bp), invE (766 bp), virF (614 bp) and sigA (430 bp) on 1.5% agarose gel. First lane: marker (100 bp), Lane 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 56, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 26, 24, 25, 59, 60, extracted DNA of Shigella isolate.
4.5. Sequencing Results
Multiplex PCR products of three genes, including virF, invE, and sigA, were purified for sequencing along with the used primers. The sequences were evaluated against all nucleotide accessions available at the NCBI GenBank DNA Database using online BLAST software. The sequences of virF, invE, and sigA have been submitted to the GenBank database with accession numbers CP055137.1, CP055125.1, and CP055125.1, respectively.
4.6. Investigating the Association Between Antibiotic Resistance Genes and the Phenotype of Antibiotic Resistance in Shigella Isolates
To investigate the association between the presence of ARGs and the phenotype of antibiotic resistance (IPM, AM, AMX, FOX, FEP), Shigella isolates were analyzed via chi-square and Cramer's V tests. Based on the results, significant and moderate associations were found between the imipenem and amoxicillin resistance phenotypes and the presence of the blaCTX-M1 gene (P < 0.05), and also between the cefepime resistance phenotype and the presence of the blaCTX-M8 gene (P = 0.06), but not with other antibiotics. The research showed that, apart from these genes, none of the other genes studied significantly correlated with antibiotic resistance. Table 5 shows the association between genes and the antibiotic resistance profile in Shigella isolates.
| Antibiotics | Chi-square | Cramer's Coefficient | P-Value |
|---|---|---|---|
| blaTEM | |||
| Imipenem | 5.48 | 0.96 | 0.760 |
| Ampicillin | 2.45 | 0.65 | 0.881 |
| Amoxicillin | 0.73 | 0.35 | 0.788 |
| Cefoxitin | 0.17 | 0.17 | 0.896 |
| Cefepime | 5.08 | 2.91 | 0.079 |
| blaOXA | |||
| Imipenem | 1.12 | 0.43 | 0.946 |
| Ampicillin | 1.36 | 1.51 | 0.506 |
| Amoxicillin | 3.90 | 0.81 | 0.533 |
| Cefoxitin | 0.92 | 0.39 | 0.761 |
| Cefepime | 1.59 | 1.63 | 0.450 |
| blaCTX-M1 | |||
| Imipenem | 2.94 | 2.22 | 0.229 |
| Ampicillin | 7.84 | 3.53 | 0.024 |
| Amoxicillin | 4.58 | 2.76 | 0.032 |
| Cefoxitin | 9.51 | 1.26 | 0.329 |
| Cefepime | 2.67 | 0.67 | 0.875 |
| blaCTX-M8 | |||
| Imipenem | 1.45 | 1.56 | 0.483 |
| Ampicillin | 1.64 | 1.65 | 0.440 |
| Amoxicillin | 2.43 | 2.02 | 0.118 |
| Cefoxitin | 1.63 | 1.65 | 0.201 |
| Cefepime | 10.31 | 4.15 | 0.006 |
5. Discussion
Shigellosis is a significant health problem affecting people worldwide, resulting in approximately 700,000 deaths yearly due to severe diarrhea. Despite improved public health measures, reports of shigellosis persist (30). Antimicrobial resistance (AMR) is a looming crisis that poses a significant threat to public health, affecting humans, animals, and the environment (31). Overuse and misuse of antibiotics have contributed to AMR. Understanding regional drug resistance patterns and continuously monitoring the involved resistance genes can help elucidate mechanisms of drug resistance in Shigella spp. and aid in the development and implementation of preventive and control measures (32).
In a study by Beladi Ghannadi et al., out of 52 Shigella isolates, more than 67% were MDR. The highest rates of resistance were observed for cephalothin, tetracycline, amikacin, trimethoprim-sulfamethoxazole, and ampicillin, while the lowest resistance rate was observed for ciprofloxacin (33). In this study, 36% of the examined isolates (n = 60) were MDR, and the highest resistance was observed for tetracycline and beta-lactam antibiotics (ampicillin and amoxicillin). Furthermore, another study in 2019 aimed to identify the drug resistance and resistance mechanisms of S. flexneri. Out of 105 S. flexneri isolates collected, 34 (32.4%) were ESBL-producing isolates. All ESBL-producing isolates were sensitive to cefoxitin and imipenem and resistant to ciprofloxacin. ESBL-producing isolates were highly resistant to ampicillin, cefotaxime, tetracycline, chloramphenicol, trimethoprim-sulfamethoxazole, ceftazidime, and cefepime (34). The resistance reported in the study by Bian et al. was higher than in the present study, especially for cephalosporins, which can be attributed to the spread of ESBL strains in the investigated isolates 34.
Our research and previous related studies have shown that Shigella isolates are resistant to amoxicillin, ampicillin, and tetracycline antibiotics, indicating that these antibiotics are no longer reliable choices for treatment. However, the observed differences in resistance to other antibiotics can be attributed to differences in the source of isolation, geographical region, and strain isolated (35-37).
Resistance to third-generation cephalosporins is a significant public health issue, especially in developing countries. This resistance is usually caused by the production of ESBL enzymes, which are often carried on plasmids (38, 39). Studies conducted in Iran found higher rates of ESBL-producing Shigella strains than have been reported in many other countries (40, 41). In this study, the presence of the six most common ESBL-producing genes (blaCTX-M1, blaCTX-M2, blaCTX-M8, blaSHV, blaTEM, and blaOXA) was investigated, and the most predominant ESBL gene was blaCTX-M1.
The results generated by our study are consistent with those of the study by Beladi Ghannadi et al. In their study, the blaTEM gene was found to be the most prevalent among Shigella isolates, followed by the blaCTX-M gene. The blaSHV gene was not detected in any of the isolates, indicating it was the least abundant (33). The blaSHV appears to have a low detection rate among Shigella isolates in Iran (42). Additionally, Toy et al. reported the prevalence of blaCTX-M1 more than other types of this gene (43). Some studies show the opposite of these results, such as a study by Hussain et al., which showed a higher prevalence of the blaCTX-M2 gene compared to our study (44), and another study by Shahin et al. from Iran, which reported contrasting results for Shigella spp. isolated from food samples (45). These differences may be due to variations in the study year, geographical location, strains, and the number of samples (46).
Generally, the results show that ESBL-producing Shigella isolates are increasing in prevalence and the beta-lactamase gene blaCTX-M is spreading rapidly. It seems that the clonal spread of strains carrying the blaCTX-M gene, the pattern of using cephalosporins, and the transfer of plasmids between different strains play an important role in the high regional prevalence of blaCTX-M genes in Enterobacteriaceae strains (47, 48).
The pathogenicity of Shigella spp. is connected to the presence of various virulence determinants, which enable the bacteria to invade and spread within the cells of the colonic epithelium (49). This study examined five virulence genes (ial, virF, invE, sigA, and pic) using a multiplex PCR assay. The distribution of virulence genes ial, virF, invE, sigA, and pic in the studied isolates was obtained. The high abundance of virF, invE, and sigA genes suggested that this classical regulatory pathway of Shigella spp. virulence gene expression might play a key role in its pathogenesis.
Additionally, the current research showed that the simultaneous presence of more than two studied virulence genes in MDR isolates was prevalent, with the pattern of invE, virF, and sigA frequently observed among MDR isolates. Across various studies conducted in Iran, a total of 667 clinical isolates were identified as MDR and extended-ESBL producers (50). Research has shown that Shigella isolates collected from water sources, like those from human sources, exhibit high levels of antibiotic resistance. For example, a study by Shahin et al. reported all Shigella isolates obtained from water samples as MDR (42). Furthermore, some MDR isolates in this research did not carry any of the studied ARGs. This result indicates that resistance to these antibiotics in these isolates could be due to other mechanisms of resistance that need further research (51).
Some studies point to a possible association between increased antibiotic resistance in bacteria and increased virulence (13, 52, 53). A conceivable explanation is that bacteria repeatedly exposed to antibiotics may become resistant to multiple drugs, which could result in the emergence of highly virulent strains (54, 55). Therefore, this result (Table 3) indicates that isolates resistant to three or more antibiotics are also likely to be more virulent, providing evidence for the hypothesis that resistance and virulence may be related.
Additional findings from this study showed the presence of blaCTX-M in many (19/22) of the MDR isolates. Similarly, several virulence genes (invE, virF, sigA, ial, and pic) were also present in the MDR isolates. However, it is important to note that not all MDR isolates carried both the studied resistance and virulence genes. In some MDR patterns, beta-lactamase genes were not detected (marked as "-ND"), and a few patterns had MDR isolates without any beta-lactamase genes. Likewise, some patterns with MDR isolates did not have all the virulence genes listed. Further examination of additional genes within this group might likely lead to a modification of the results.
The current study is limited by the small sample size, which could affect the generalizability of the findings. Additionally, the study focused on Tehran, which may not be representative of the entire country. Information on patient clinical characteristics and treatment history that could lead to a clearer interpretation of antibiotic resistance patterns was also not available. Considering the high prevalence of multidrug resistance observed in this study, it is necessary to establish a continuous monitoring system to track the trend of antibiotic resistance in Shigella isolates in Iran.
5.1. Conclusions
In this study, high resistance to beta-lactams and tetracycline, as well as a high prevalence of blaCTX-M genes, were observed. Furthermore, high resistance to the first-line treatment of shigellosis, i.e., ampicillin, was also noted. With these limitations in treatment options and the emergence of cephalosporin-resistant strains, new strategies for treating shigellosis need to be developed, with a focus on continuous surveillance. This will help us develop more effective treatments and control strategies.