1. Background
The opportunistic pathogen Acinetobacter baumannii is classified within the A. calcoaceticus-A. baumannii complex, along with other phenotypically similar Acinetobacter species (A. calcoaceticus, A. Pitti, A. Seifert, A. djikshoorniae, and A. nosocomial), which are difficult to distinguish from each other without genomic typing (1). Acinetobacter baumannii is frequently isolated from devices, health workers, and patients in the hospital environment and causes significant and health-threatening nosocomial infections and infections are usually observed in patients treated in intensive care units (ICUs) and burn units worldwide, including in our country (2, 3). The fact that A. baumannii is both inherently resistant and easily becomes resistant to antibiotics is why it is frequently reported in hospitals around the world in relation to epidemics and classified among the six most dangerous pathogens (4, 5). Its current resistance to commonly used antibiotics, including carbapenems, fluoroquinolones, aminoglycosides, and tetracyclines, has raised important health concerns, such as increased length of hospitalization and higher mortality rates among infected patients (1, 6).
Antibiotic resistance in A. baumannii is caused by mechanisms involving decreased expression of outer membrane proteins and/or the presence of efflux pumps. However, carbapenem resistance is specifically caused by the presence or enhanced expression of beta-lactamase enzymes (7, 8). These enzymes are divided into classes A, B, C, and D according to the Ambler classification as previously reported (7, 9, 10). Mobile genetic elements, such as plasmids, transposons, and integrons, can contain resistance-causing beta-lactamase gene regions and are important factors in the spread of resistance in A. baumannii strains (11).
The ISAba-I insertion element is preceded by gene regions that encode beta-lactamase genes, and it increases their expression by acting as a promoter for the co-existing beta-lactamase genes. This genetic flexibility gives A. baumannii an unpredictable resistance profile, so understanding the various modes of resistance is of great importance for the prevention of spread and for the treatment of hospitalized patients (12). Studies carried out in our country have indicated increasing levels of antibiotic resistance in Acinetobacter species, with susceptibility ratios differing due to different epidemiological conditions and antibiotic use strategies (13). Studies from many regions of our country have shown drug resistance profiles related to A. baumannii, and mechanisms that induce resistance have been proposed; however, no data has been reported for Mugla province (14).
2. Objectives
The aims of the present study were, therefore, to identify the gene regions that encode beta-lactamase enzymes classified in Ambler class B and D and clonal relatedness among multi-drug resistant (MDR) A. baumannii strains isolated in two different intervals, including September 2011-June 2012 and the year 2019.
3. Methods
3.1. Bacterial Isolates and Antibiotic Susceptibility Tests
The study investigated 96 non-duplicate A. baumannii isolates, including 61 collected between September 2011 and June 2012 and 35 collected in 2019. Samples were taken from hospitalized patients in a 650-bed tertiary care at Mugla Sitki Kocman University Education Research Hospital, Mugla, Turkey, and examined at the Medical Microbiology Laboratory. In addition to conventional identification methods, the isolates were identified using API20 NE (bioMérieux, France) and BD Phoenix (U.S.A.) systems. Each isolate was divided into multiple aliquots for long-term storage at -80°C in brain-heart infusion (Merck) broth containing 30% glycerol and an aliquot used only once.
This multi-storage process protects against the loss of resistance genes and minimizes degradation caused by freeze-thawing. Antibiotic susceptibility tests were performed using the disk Kirby-Bauer diffusion method on Mueller-Hinton agar. For the identification and antibiotic susceptibility testing of the 2019 isolates, the BD Phoenix automated system was used and interpreted according to EUCAST version 9.0 (15). The minimum inhibitory concentrations (MIC) of imipenem and colistin for all isolates were determined by microdilution method using cation-added Mueller-Hinton broth (Merck). A. baumannii ATCC 19606 was used as the standard strain.
3.2. DNA Isolation and Detection of Resistance Genes
Total bacterial DNA was obtained using a DNA isolation kit (Fermentas, Finland) in accordance with the manufacturer's recommendations and used for the detection of resistance genes by a conventional polymerase chain reaction (PCR) method. Detection of resistance genes was carried out in Mugla Sitki Kocman University, Faculty of Health Sciences Research Laboratories. The stock isolates were seeded on eosin methylene blue (EMB) agar (Merck) and incubated overnight at 36°C. Total bacterial DNA was extracted with a DNA isolation kit and stored at -20°C until used in the assay.
The presence of gene regions encoding the beta-lactamase enzymes blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, blaNDM, blaIMP, and blaVIM, as well as the ISAba-I insertion element, were investigated in the total genomic DNA. The position of ISAba-I in relation to the OXA carbapenemase genes was determined by combinations of forward primers designed for ISAba-I and reverse primers for OXA gene regions. The primers used in the conventional PCR method for the amplification of gene regions (12, 16, 17) are presented in Table 1. The following PCR mixture was used for genomic DNA for each isolate: 0.2 L Taq DNA polymerase (5 U/L), 2.5 L dNTP (2 mM each), 2 L MgCl2 (25 mM), 0.5 L primer (20 pmol/L), 2.5 L 10× Taq DNA polymerase buffer (Thermo Fisher Scientific, U.S.A.), and 3 L DNA in a final volume of 25 L.
The PCR procedure was performed in 30 cycles in a thermal cycler (Techne, Model 4000, UK), as follows: Initial denaturation 5 min at 94°C; denaturation 50 s at 94°C, annealing for 50 s at the temperatures presented in Table 1 for each primer, extension 50 s at 72°C, and a final extension step for 7 min at 72°C. The PCR products were subjected to 2% agarose gels containing 0.5% ethidium bromide. All amplicons were analyzed using 100 bp and 1 Kb DNA size ladder (Thermoscientific, Lithuania). The bands were visualized with a UV imaging device (Vilber Lourmart, France) and analyzed with the E-capt software (Vilber Lourmart, France) supplied with the device.
| Gene Regions | 5’ → 3’ Primer Pairs | T (C) | References |
|---|---|---|---|
| blaOXA-23-like | F- GAT CGG ATT GGA GAA CCA GA | 57 | (16) |
| R- ATT TCT GAC CGC ATT TCC AT | |||
| blaOXA-24-like | F- GGT TAG TTG GCC CCC TTA AA | 58 | (16) |
| R- AGT TGA GCG AAA AGG GGA TT | |||
| blaOXA-51-like | F- TAA TGC TTT GAT CGG CCT TG | 57 | (16) |
| R- TGG ATT GCA CTT CAT CTT GG | |||
| blaOXA-58-like | F- AAG TAT TGG GGC TTG TGC TG | 58 | (16) |
| R- CCC CTC TGC GCT CTA CAT AC | |||
| blaNDM | F- GCA GGT TGA TCT CCT GCT TG | 60 | (17) |
| R- ACG GTT TGG CGA TCT GGT | |||
| blaIMP | F- CTA CCG CAG CAG AGT CTT TGC | 60 | (17) |
| R- GAA CAA CCA GTT TTG CCT TAC C | |||
| blaVIM | F- GAT GGT GTT TGG TCG CAT A | 60 | (17) |
| R- CGA ATG CGC AGC ACC AG | |||
| ISAba-I | F- CAC GAA TGC AGA AGT TG | 52 | (12) |
| R- CGA CGA ATA CTA TGA CAC | |||
| ISAba-I-blaOXA-23 | F- CAC GAA TGC AGA AGT TG | 60 | (12) |
| R- ATT TCT GAC CGC ATT TCC AT | |||
| ISAba-I-blaOXA-24 | F- CAC GAA TGC AGA AGT TG | 57 | (12) |
| R- AGT TGA GCG AAA AGG GGA TT | |||
| ISAba-I-blaOXA-51 | F- CAC GAA TGC AGA AGT TG | 53 | (12) |
| R- TGG ATT GCA CTT CAT CTT GG | |||
| ISAba-I-blaOXA-58 | F- CAC GAA TGC AGA AGT TG | 60 | (12) |
| R- CCC CTC TGC GCT CTA CAT AC |
Abbreviation: T, annealing temperature.
3.3. Repetitive Extragenic Palindromic PCR
The repetitive extragenic palindromic (REP) like elements in the genomic DNA extracted from A. baumannii isolates were amplified with the primer pair REP1 5′-IIIGCGCCGICATCAGGC-3′ and REP2 5′-ACGTCTTATCAGGCCTAC-3′ as described previously (18). Repetitive extragenic palindromic PCR was performed in a final volume of 25 μL, including 0.5 μL of each forward and reverse primers and 3 μL of the template DNA. Amplification was carried out with an initial denaturation at 95°C for 10 min, followed by 30 cycles of denaturation (95°C, 1 min), annealing (40°C, 1 min), extension (72°C, 2 min) and a final extension at 72°C for 15 min. 15 μL of PCR products were loaded in 1.5% agarose gel for electrophoresis. The REP-PCR profiles were analyzed with BioNumerics version 7.6 software to demonstrate clonal distribution (Applied Maths, Belgium). Cluster analysis was arranged using an unweighted pair-group method using arithmetic averages (UPGMA) and a Dice similarity coefficient with 1.5% tolerance. Eighty five percent similarity was considered as the cut-off value for pertaining to the same cluster.
3.4. Data Analysis
The data were analyzed using Mann-Whitney U parametric and non-parametric tests and the chi-square test. P-Value < 0.05 was accepted as statistically significant. IBM SPSS Statistics 21 statistical software was used for data analysis.
4. Results
In total, 96 MDR A. baumannii clinical isolates were included in the study; 61 were obtained between September 2011 and June 2012, and the remaining 35 were collected in 2019. A list of the clinical samples and the services from which the 96 A. baumannii isolates were obtained is presented in Table 2. In total, 80 (83.3%) clinical samples were taken from the respiratory tract; most of these were tracheal aspirates (n = 62; 64.6%), followed by sputum (n = 11; 11.4%). The distribution of clinical samples between two different specimen collection periods showed no statistically significant difference.
| Specimen | 2011 - 2012 (n = 61) | 2019 (n = 35) | Total (n = 96) |
|---|---|---|---|
| Tracheal aspirate | 39 (63.9) | 23 (65.7) | 62 (64,6) |
| Sputum | 8 (13.1) | 3 (8.6) | 11 (11,4) |
| Urine | 6 (9.9) | 3 (8.6) | 9 (9.4) |
| Throat | 6 (9.9) | 1 (2.8) | 7 (7.3) |
| Blood | 1 (1.6) | 3 (8.6) | 4 (4.2) |
| Wound | 1 (1.6) | 2 (5.7) | 3 (3.1) |
| Total | 61 (100.0) | 35 (100.0) | 96 (100.0) |
| Units | |||
| Internal Med. ICU | 16 (26.2) | 8 (22.9) | 24 (25.0) |
| Anesthesia ICU | 12 (19.7) | 8 (22.9) | 20 (20.8) |
| Neurology ICU | 5 (8.2) | 7 (20.0) | 12 (12.5) |
| Surgery ICU | 8 (13.1) | 6 (17.1) | 14 (15.6) |
| Pulmonary Dis. Unit. | 11 (18.0) | 0 | 11 (11.4) |
| Surgery Unit. | 6 (9.9) | 0 | 6 (6.2) |
| Internal Med.Unit. | 3 (4.9) | 1 (2.8) | 4 (4.2) |
| Others b | 0 | 5 (14.3) | 5 (5.2) |
| Total | 61 (100.0) | 35 (100.0) | 96 (100.0) |
Abbreviation: ICU: Intensive care unit.
a Values are expressed as No. (%).
b Other units are; emergency: 2; urology: 1; cardiovascular surgery ICU: 1; infection: 1.
Overall, 71 (74.0%) of the total MDR isolates were obtained from ICUs; 41 (67.2%) of the isolates were from 2011 - 2012 and 30 (85.7%) were from 2019 (Table 2). The ICUs where the isolates were obtained were internal medicine (n = 24; 25.0%), anesthesia and reanimation (n = 20; 20.8%), neurology (n = 12; 12.5%), surgery (n = 14; 15.6%), and cardiovascular surgery (n = 1; 1.0%). The antibiotic resistance patterns and gene regions encoding beta-lactamase enzymes determined for the isolates are summarized in Table 3. The disk diffusion tests or automated system analysis confirmed that all isolates were resistant to ciprofloxacin, piperacillin, piperacillin-tazobactam, imipenem, and meropenem. The resistance rate for gentamicin was detected as 96.7% (n=59) in 2011 - 2012 isolates and for trimethoprim-sulfamethoxazole as 91.4% (n = 32) in 2019 isolates. Resistance ratios were determined as 100% (n = 96) and 21.9% (n = 21) for imipenem and colistin, respectively, by the MIC method (Table 3). Colistin resistance was detected in 6.6% (n = 4) of the 2011–2012 isolates but in 48.6% (n = 17) of the 2019 isolates, confirming an increase in the number of colistin-resistant bacteria over the years (P = 0.000).
| Variables | 2011 - 2012 (n = 61) | 2019 (n = 35) | Total (n = 96) |
|---|---|---|---|
| Antibiotic resistance ratios | |||
| Amikacin | 59 (96.7) | 33 (94.3) | 92 (95.8) |
| Gentamicin | 59 (96.7) | 35 (100) | 94 (97.9) |
| Ciprofloxacin | 61 (100) | 35(100) | 96 (100) |
| Piperacillin | 61 (100) | 35 (100) | 96 (100) |
| Piperacillin-tazobactam | 61 (100) | 35(100) | 96 (100) |
| Trimethoprim-sulfamethoxazole | 61 (100) | 32 (91.4) | 93 (96.9) |
| Meropenem | 61 (100) | 35 (100) | 96 (100) |
| Imipenem (MIC method) (> 4 mg/L) | 61 (100) b | 35 (100) b | 96 (100) |
| Colistin (MIC method) (> 2 mg/L) | 4 (6.6) c | 17 (48.6) c | 21 (21.9) |
| Gene regions | |||
| blaOXA-23-like | 61 (100) | 32 (91.4) | 93 (96.9) |
| blaOXA-24-like | 0 | 3 (8.6) | 3 (3.1) |
| blaOXA-51-like | 61(100) | 35 (100) | 96 (100) |
| blaNDM | 2 (3.3) | 3 (8.6) | 5 (5.2) |
| ISAbaI | 61 (100) | 35 (100) | 96 (100) |
| ISAbaI/blaOXA-23-like | 51 (83.6) | 31 (88.6) | 83 (86.5) |
| ISAbaI/blaOXA-51-like | 1 (1.6) | 1 (2.9) | 2 (2.1) |
| blaOXA-23-like/blaOXA-24-like | - | 2 (5.7) | 2 (2.1) |
| blaOXA-23-like/ ISAbaI-blaOXA-51-like | 1 (1.6) | 1 (2.9) | 2 (2.1) |
| blaOXA-24-like/ ISAbaI-blaOXA-23-like | - | 1 (2.9) | 1 (1.1) |
a Values are expressed as No. (%).
b P = 0.006; imipenem MIC 2011-2012 vs. 2019;
c P = 0.000; colistin MIC 2011-2012 vs.2019.
The MIC values for imipenem in 2011 - 2012 isolates vary between 8 - 128 mg/L; the most prevalent was 16 mg/L for 40 of 61 isolates. For 2019 isolates, it was 32 mg/L for 20 of 35 isolates. As summarized in Table 4, there is a statistically significant increase (P = 0.006) between the two periods. On the other hand, colistin MIC values were between 4 - 16 mg/L for 4 of 61 isolates in the 2011 - 2012 period, but in the year 2019, the MIC range was raised to 64 mg/L for 14 of 35 isolates (P = 0.000). When compared to the reference periods, a statistically significant increase in MIC values was noted for the MDR A. baumannii isolates (Table 4).
| Variables | 2011 - 2012 (N) | 2019 (N) | P-Value |
|---|---|---|---|
| Imipenem MIC (mg/L) | 0.006 | ||
| 8 | 11 | 3 | |
| 16 | 40 | 8 | |
| 32 | 8 | 20 | |
| 64 | 1 | 2 | |
| 128 | 1 | 2 | |
| Total | 61 | 35 | |
| Colistin MIC (mg/L) | 0.000 | ||
| 4 | 2 | 1 | |
| 8 | 1 | - | |
| 16 | 1 | - | |
| 32 | - | 2 | |
| 64 | - | 14 | |
| Total | 4 | 17 |
Abbreviation: N, number of resistant isolates; MIC, minimum inhibitory concentrations.
The distribution of beta-lactamase genes that cause antibiotic resistance is summarized in Table 3. The blaOXA-51-like gene region was present (100%) in all MDR A. baumannii isolates, as was the ISAba-I insertion element. The blaOXA-24-like gene region was detected in only 3 of the isolates from 2019, and it co-occurred with blaOXA-23-like in 2 of them. All the MDR A. baumannii isolates from 2011 - 2012 showed the presence of blaOXA-23-like, whereas 32 (91.4%) of the 2019 isolates showed the presence of blaOXA-23-like. The blaNDM gene region, which encodes the metallobeta-lactamase enzyme, was detected in a total of 5 (5.2%) isolates; 2 (3.3%) from 2011 - 2012 and 3 (8.6%) from 2019. However, none of the 96 isolates had a positive PCR result for the blaOXA-58-like, blaIMP, or blaVIM beta-lactamase encoding genes.
The ISAbaI insertion element was found upstream of the blaOXA-23-like genes in 83 (86.5%) of the total isolates, 51 (83.6%) from 2011 - 2012, and 31 (88.6%) from 2019. ISAba-I /blaOXA-51-like co-occurred in 2 isolates (2.1%), one from each collection period.
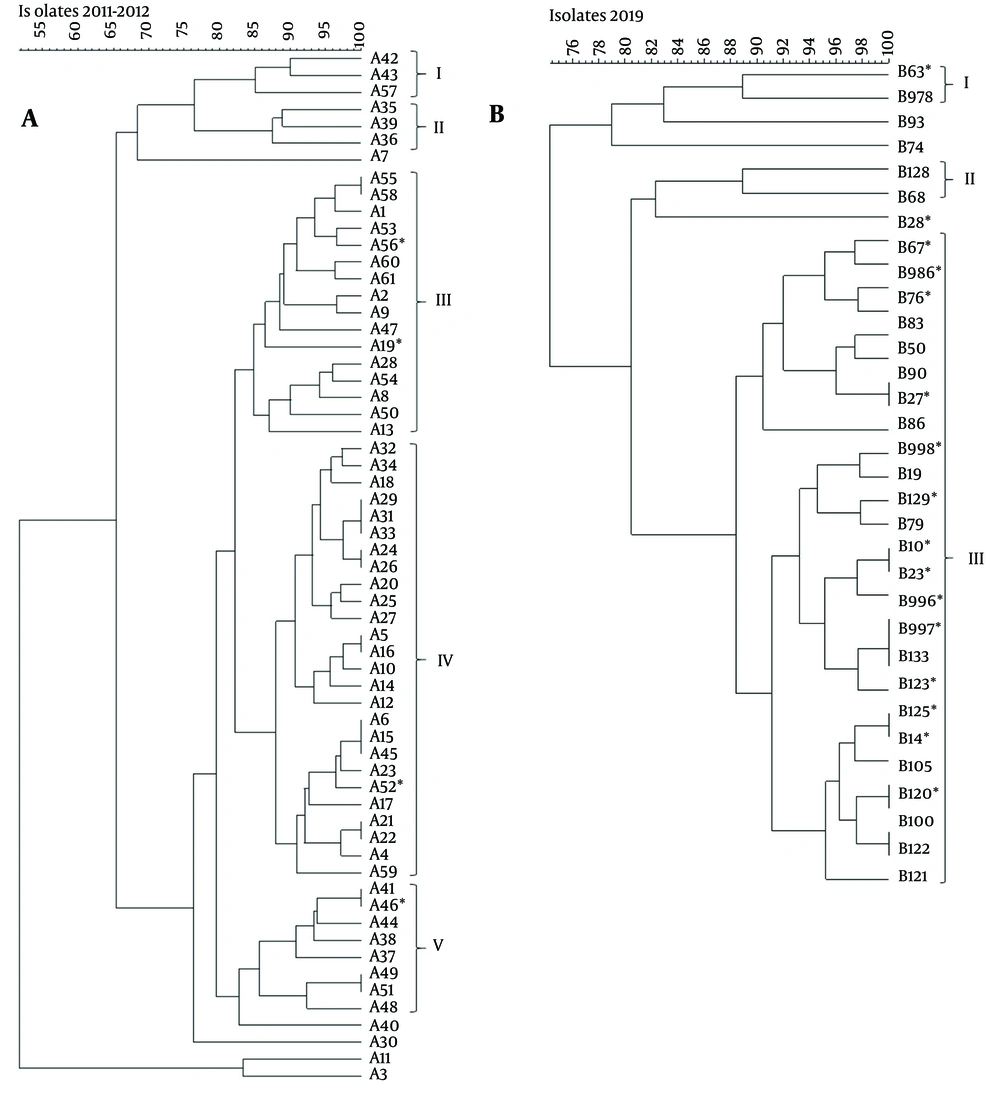
Repetitive extragenic palindromic-PCR was performed in order to demonstrate the clonality of the 2011 - 2012 and 2019 isolates separately (Figure 1 A and B). The cluster analysis of REP-PCR products of the 2011 - 2012 isolates indicated epidemiologically related isolates distributed through five clusters, while A3, A7, A11, A30, and A40 were considered as unrelated to any cluster (Figure 1 A). Among these five clusters, 18 (29.5%) isolates were demonstrated genetically as indistinguishable or closely related according to Tenover et al. criteria (19). The largest cluster identified was cluster IV, containing 26 (42.6%) isolates. Four colistin-resistant isolates were identified and distributed as A19 and A56 were in cluster III, A52 was in cluster IV, and A46 was in cluster V. On the other hand, as summarized in Figure 1 B, 35 isolates from 2019 were distributed through three clusters, whereas 3 of the isolates (B28, B74, and B93) could not be identified within the 85% similarity cut-off value and were not incorporated in any cluster. 28 of 35 (80%) isolates existed in cluster III and demonstrated genetically as indistinguishable or closely related with similar REP-PCR patterns above 85% similarity. Among 17 colistin-resistant isolates, 15 of them existed within the largest cluster III.
A-B, dendogram representing repetitive extragenic palindromic-PCR profile analysis of genomic DNA from multi-drug resistant (MDR) Acinetobacter baumannii isolates. The asterisk indicates the colistin-resistant A.baumannii isolates. The cut-off value was set at 85% for the similarity coefficient with 1.5% tolerance. The identification number of the isolates was presented throughout the profiles. A, represents the REP-PCR profiles of 61 MDR isolates, including four colistin-resistant strains, collected in 2011 – 2012; and B, demonstrates 35 MDR A.baumannii, including 17 colistin-resistant isolates collected in 2019.
5. Discussion
Acinetobacter baumannii isolates, which are resistant to most known antibiotics, can persist in the hospital environment for prolonged periods and cause hospital infections (12). The greatest problem arising from A. baumannii infections is difficulty in treatment due to their multidrug resistance (6). Various mechanisms have been proposed to cause resistance to beta-lactam antibiotics, such as beta-lactamase enzyme production, changes in external membrane proteins, expression of proteins that bind penicillin, and the presence or increased activity of drug efflux pumps (20, 21). Mortality rates between 8% and 43% are reported for MDR A. baumannii infections (7).
Predisposing factors for Acinetobacter spp. infections include antibiotic therapy, major surgery, burns, immune suppression, and especially mechanical ventilation. More than 30% of the clinical isolates of Acinetobacter spp. detected in ICUs were resistant to at least three classes of antibiotics, most frequently third-generation cephalosporins, fluoroquinolones, and carbapenems (13). Previous studies have shown that more than 96% of A. baumannii strains, especially those isolated from hospital environments, were resistant to all tested antimicrobial agents except colistin. More than 60% of the clinical specimens were taken from respiratory tract infections of patients in intensive care or burn units, and the majority of these infected patients were thought to be immunocompromised and/or had an underlying chronic illness (6, 17). Our findings agreed with these previous studies, as 83.3% (n = 80) of our isolates were obtained from respiratory-related samples, and 74.0% (n = 71) were obtained from immunocompromised ICU patients who were presumed susceptible to opportunistic infectious agents.
In recent years, the hospital environment has been mentioned as a prime source of MDR A. baumannii infections and transmissions in many regions of the world. The most important issue for consideration under these conditions is that not many antimicrobial treatment options remain available for the treatment of drug-resistant phenotypes (6). The fact that the MDR isolates included in the study were 96.9% resistant to amikacin and gentamycin and 100% resistant to all other antibiotics (except colistin) is proof that these isolates were MDR strains. The resistance to colistin varies among the studies; for example, 0.6% resistance was found in the multicenter research conducted by Beriş et al. in Turkey in 2016 (13). By contrast, the 2016 CAESAR report for Turkey indicated a ratio of MDR A. baumannii isolates of 83.5% and a resistance to colistin of 4% (14). A multicenter study of 10 hospitals reported a 99.4% resistance rate for imipenem in MDR A. baumannii isolates and a 1.2% resistance to colistin (22). In our study, the resistance rate for colistin was 6.6% (n = 4) for the isolates from 2011 - 2012 but 48.6% (n = 17) for the isolates from 2019, indicating a substantial increase in resistance rates and MIC values for colistin in the MDR isolates over the years. The resistance rate for all 96 isolates was 21.9% (n = 21) for colistin.
Carbapenem resistance has risen to over 80% in recent years from its initial reported rate of 1.3% (8). Notably, all isolates resistant to carbapenem are MDR or pan-drug resistant strains. In their meta-analysis, Bialvaei et al. reported that 67% of the isolates in Iraq, 85% in Kuwait, and 83% in the Arab Emirates were MDR A. baumannii (11). In 2013, Çiftçi et al. (8) reported higher rates of meropenem resistance than imipenem resistance in their multicenter study. They also stated that the difference observed in the resistance rates to meropenem and imipenem has become smaller over the years (8). In our study, all the isolates evaluated in both periods were resistant to meropenem and imipenem.
The MIC values for imipenem for isolates from 2011 - 2012 and from 2019 also increased (P = 0.006), in agreement with other ratios reported in Turkey (8). The spread of MDR A. baumannii isolates is often clonal, and the clones show genetic homology with each other (5, 13). Integrons harboring bla genes, which encode multiple antibiotic resistances, play a significant role in dissemination as natural cloning systems and expression vectors (23). However, the masses of genes that encode beta-lactamase enzymes (blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and blaOXA-58-like) and their distribution vary by country. The blaOXA-51-like gene is unique to A. baumannii and occurs all over the world simply because it is naturally found in this bacterium. By contrast, blaOXA-23-like is widespread in Europe and the Far East, while blaOXA-24-like exists in Western Europe and America, and the blaOXA-58-like gene has been reported in several countries, including Turkey (8, 13, 24).
The blaOXA-51-like and blaOXA-23-like genes play a major role in carbapenem resistance, as all resistant A. baumannii isolates contain the blaOXA-51-like gene region, and up to 98.4% of MDR isolates contain the blaOXA-23-like gene (22). In previous years, the presence of the blaOXA-58-like gene region was reported at a ratio of 17.1%, but its presence has been reported less frequently, at 6%, in recent years. One possible cause of this decrease may be the widespread colonization of blaOXA-23-like positive isolates in ICUs or throughout hospital services, so this gene region is now the dominant gene cluster. Lately, the blaOXA-24-like gene region has been reported at an average rate of 10% in Turkey (5, 8, 24). Similarly, Abbasi et al. detected the presence of blaOXA-23-like .
In all their MDR A. baumannii isolates but could not detect blaOXA-58-like in any isolate (6). Two different studies were performed with carbapenem-resistant A. baumannii in China. Wang et al. found high carrying rates of blaOXA-51-like (66.9%), and blaOXA-23-like (74.5%) genes (23), and Huang et al. found to be 90% and 77.5%, respectively (25), which were more common in China. Wang et al. also reported lower rates of blaNDM, blaOXA-24-like, and blaOXA-58-like genes (23). In our study, the blaOXA-51-like and blaOXA-23-like gene regions were detected in all (100%) of our MDR A. baumannii isolates, and blaOXA-24-like was present in 3 of the isolates obtained in 2019 (3.1%). The blaOXA-58-like gene region was not detected in any of our isolates. A comparison of our patient groups for the two research periods revealed a noticeable increase in MIC values for the MDR A. baumannii isolates containing the blaOXA-23-like gene region over the years.
The blaNDM, blaIMP, and blaVIM gene regions are not widely reported in Turkey. One study performed in 2016 showed the presence of blaNDM in 22% (n = 12) of 55 A. baumannii isolates in Turkey (26). Although blaIMP and blaVIM have been detected in the Far East and Europe, the data in Turkey indicate very limited or no distribution (22, 26). A multicenter study conducted by Boral et al. reported that 96% of 164 A. baumannii isolates contained the blaOXA-23-like gene region, and 3% contained the blaOXA-58-like gene region; however, blaOXA-24-like, blaNDM, and blaIMP were not detected (22). In their systematic review of 12 years of data, Kahraman Kilbas et al. reported that the presence of blaOXA-58-like is more common in the east of Turkey, while the presence of blaOXA-23-like is more common in the west. Additionally, they reported that the prevalence of blaNDM positivity is not very common, with a ratio of 1.1% (27). In this study, blaNDM was detected in 5 (5.2%) of the 96 MDR A. baumannii isolates, 2 from 2011 - 2012 (3.3%) and 3 (8.6%) from 2019, but blaIMP and blaVIM were not detected.
In parallel with the findings of our study showing a high presence of blaOXA-51-like and blaOXA-23-like, Lowe et al. (28) found the positivity rates of these genes to be 100% and 96%, respectively, in their research on MDR A. baumannii. Additionally, they reported the presence of the ISAba-I insertion element to be 100%, similar to our results. However, contrary to our findings, they did not observe any isolate that carried the ISAba-I element upstream of the blaOXA-23-like gene (28). A previous study of the ISAba-I insertion element showed its co-occurrence with the blaOXA-23-like gene region in 97% of carbapenem-resistant isolates (7). Similarly, Abbasi et al. (6) found a co-occurrence of ISAba-I and blaOXA-23-like presence in 80% of the MDR A. baumannii isolates obtained over a one-year period and confirmed that all isolates were resistant to carbapenems. The ISAba-I/blaOXA-51-like positivity was reported as 65% in their isolates (6). We found ISAba-I/blaOXA-23-like positivity in 86.5% of the MDR isolates from both collection periods, similar to these previous findings. However, the ISAba-I/blaOXA-51-like positivity was observed in only 2 (2.1%) of 96 isolates. We found no statistically significant relation between the presence of beta-lactamase enzyme-encoding genes and colistin and imipenem MIC values.
Huang et al., investigating clonal relationships using the PFGE method, indicated that multidrug-resistant isolates clustered within the same group in their results (25). Similarly, in our study, it was found that colistin-resistant A. baumannii isolates, particularly those obtained in 2019, with high carrying rates of blaOXA-51-like (100%) and blaOXA-23-like (91.4%), and to a lesser extent blaOXA-24-like (8.6%) and blaNDM (8.6%) mainly clustered within cluster III. Apart from these, 2011 - 2012 isolates were distributed in five different clusters with lower rates of colistin resistance.
5.1. Conclusions
The antibiotic resistance rates in Mugla province were especially high in patients infected with A. baumannii in ICUs who underwent invasive intervention, emphasizing the importance of developing new strategies for treatment and prevention. These findings suggest epidemic threats should not be ignored in healthcare facilities, likely in this study, as depicted in Figure 1.
When both periods are considered, all 96 isolates were identified as MDR, and 21 of them were found to be colistin-resistant, of which 17 were isolated in 2019. BlaOXA-23-like was detected in 96.9% of the isolates and is the most commonly found beta-lactamase enzyme coding gene in Turkey. All the isolates have ISAba-I insertion element, and 86.5% of the isolates carried upstream of the blaOXA-23-like gene. The high coexistence of resistance coding genes observed, the increase in colistin and imipenem MIC values, and the fact that 80% of the isolates were in the same cluster of the isolates obtained in 2019 suggested that carbapenem resistance genes were frequently disseminated over an eight-year interval. We believe that elucidating and comparing the confirmed data of the research contributes significantly to the existing database, can be strengthened by further studies, and can guide the correct use of effective antibiotics.