1. Background
Hepatocellular carcinoma (HCC) accounts for a significant proportion of primary liver cancer (LC) cases (1) and is associated with high mortality worldwide (2). The incidence of HCC has been rising globally. Hepatitis B virus (HBV) infection is the most significant cause of LC, with about half of all LC cases attributable to HBV infection. HBV, a double-stranded, loosely circular DNA virus, replicates in the host through reverse transcription. Upon infecting hepatocytes, part of the viral DNA is transported to the host nucleus and converted into covalently closed circular DNA (3). Recent advances have been made in the clinical diagnosis and treatment of HCC, encompassing conventional methods such as surgery, radiotherapy, and chemotherapy, as well as biotherapy and traditional Chinese medicine. However, many patients with HCC are diagnosed at an advanced stage, the pathogenesis remains poorly understood, and targeted therapies are limited. Consequently, there has been no significant improvement in overall survival for patients with HCC (4). Additionally, studying the regulatory mechanisms of HBV is crucial for developing targeted therapies.
MicroRNA (miRNA) is a class of small non-coding RNAs that regulate genes. MicroRNAs involved in the development of LC can serve as new biomarkers for HCC. Some miRNAs can affect HBV replication directly by targeting HBV mRNA or indirectly by regulating host factors (5-9). miR-642a-5p, a newly discovered tumor-associated miRNA, has been found to be downregulated in colon cancer tissue (10) and Hodgkin lymphoma (11). Additionally, aberrant expression of miR-642a-5p is observed in multiple human cancers including lung cancer, cervical cancer, and myeloma (12). However, the mechanism of miR-642a-5p in HBV-infected LC remains unclear.
DNA Damage Inducible Transcript 4 (DDIT4), also known as DNA damage-stimulating transcript 4, is a tumor suppression protein discovered in 2002 (13). It can be stimulated under various stress conditions such as oxidative stress, endoplasmic reticulum stress, hypoxia, and heat shock. Under normal circumstances, DDIT4 is generally downregulated in major adult tissues (14), but its expression is elevated in a variety of malignant tumors. Studies have shown that elevation of DDIT4 enhances the proliferation of gastric epithelial cells (15), and DDIT4 facilitates gastric cancer proliferation and tumorigenesis via p53 and MAPK pathways (16). Moreover, in HCC patients, DDIT4 is elevated compared to healthy individuals, making it a potential biomarker for HCC (17). Nevertheless, the regulatory mechanism of DDIT4 in HBV-infected HCC remains unexplored.
2. Objectives
This study was to explore the mechanism of inhibition of HBV HCC by miR-642a-5p targeting DDIT4.
3. Methods
3.1. Sample Collection
From 2016 to 2018, 49 HBV-positive [HBV (+), pathologically confirmed] HCC tissues and paired para-cancer normal liver tissues (Normal) were collected from The Third People's Hospital of Yunnan Province. Inclusion criteria included: (1) All subjects meeting the "Clinical Diagnosis and Staging Criteria of Primary Liver Cancer" for primary LC, confirmed by pathological biopsy results; (2) an HBV DNA level exceeding 1 × 10³ copies/mL; (3) positive results for hepatitis B surface antigen (HBsAg) or HBV DNA (more than 6 months). None of the patients had received radiotherapy, chemotherapy, systemic treatment, or immunotherapy previously. The clinical stage classification of each LC adhered to the standards set by the International Union for Cancer Control, and tissue sample examinations were performed by a trained pathologist. Serum was collected from healthy volunteers (control group) and patients with acute HBV infection (acute infection group) at Guangzhou Infectious Disease Hospital, with 20 cases in each group.
3.2. Cell Culture and Screening
The human normal liver cell line HL-7702 and the stable HBV-positive cell line HepG2.2.15 (RRID: CVCL_L855) were acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The HepG2.2.15 cell line, derived from HepG2 cells transfected with a complete HBV DNA recombinant plasmid, can stably replicate and express HBV. HL-7702 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (GIBCO BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin. Both HepG2.2.15 and HL-7702 cells were cultured under identical conditions, with the addition of 2 mM L-glutamine and 200 μg/mL G418 (Invitrogen, Carlsbad, CA). The cells were maintained in a humidified incubator and passaged when the cell density reached nearly 90%.
3.3. Experimental Grouping and Cell Transfection
HL-7702 and HepG2.2.15 cells were cultured until they reached the logarithmic growth phase. The cells were then divided into seven groups and transfected with various agents: miR-642a-5p mimic, mimic-NC (negative control), miR-642a-5p inhibitor, short hairpin RNA plasmid targeting DDIT4 (sh-DDIT4), sh-NC (negative control), inhibitor-miR-642a-5p + sh-NC, and inhibitor-miR-642a-5p + sh-DDIT4, respectively. Transfections were carried out using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) or antisense oligonucleotides (ASOs). Forty-eight hours post-transfection, cells were harvested for subsequent experiments, and the efficacy of transfection was verified. To ensure the reliability of the data and the absence of bias, all subsequent cell experiments were repeated five times. The miR-642a-5p mimic, mimic-NC, miR-642a-5p inhibitor, sh-NC, and DDIT4 vector plasmids were sourced from RiboBio (Guangzhou, China).
3.4. Hepatitis B Virus DNA Detection
After 48 hours of transfection, HBV DNA was extracted from the cell culture supernatant using the QiAamp DNA Blood Mini Kit (Qiagen). Reverse transcription quantitative polymerase chain reaction for HBV DNA was conducted using a PCR fluorescence quantitative detection kit (Bioer, Hangzhou, China). The average threshold value was employed to determine the concentration of HBV DNA. The HBV DNA inhibition rate (%) was calculated using the formula: [(Number of DNA copies in the control group - number of DNA copies in the experimental group) / number of DNA copies in the control group] × 100%.
3.5. Cell Counting Kit (CCK)-8 Assay
HepG2.2.15 cells were seeded in 96-well plates (2 × 105 cells/well) and treated with 10 mL of CCK-8 (Dojindo, Japan) in each well before and after transfection at 24, 48, and 72 hours (18). Cell proliferation was measured by the optical density at 450 nm using a microplate reader (Bio-Rad) (19).
3.6. Flow Cytometry
HepG2.2.15 cells were seeded in a 6-well plate at a density of 3 × 106 cells per well and cultured for 24 hours before transfection. After 48 hours of culture in starvation conditions, double staining was performed using Annexin V/7AAD (Biolegend). The proportion of apoptotic cells was calculated, and the results were analyzed using FlowJo version 6.2 software (20).
3.7. Cell Migration and Invasion
Cells were digested with trypsin to prepare a single-cell suspension. Cell migration analysis was conducted using 24-well plates with an 8 µm pore size polycarbonate membrane. HepG2.2.15 cells were seeded in a Transwell chamber in 400 µL of serum-free DMEM at a density of 3 × 104 cells per well. Concurrently, 800 µL of medium containing 20% FBS was added to the lower chamber. After 24 hours of culture, cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Cells that had migrated through the membrane were observed under a microscope and counted. The invasion of HepG2.2.15 cells was further analyzed using a Transwell chamber assay, with the upper chamber coated with BD Matrigel™ Basement Membrane Matrix (21).
3.8. Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from HepG2.2.15 cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Quantitative RT-PCR was conducted on the StepOnePlus real-time PCR system (Applied Biosystems, Waltham, MA, USA), following the manufacturer's instructions. Reverse transcription of 5 μg of total RNA into cDNA was performed using miRNA-specific reverse transcriptase primers and the first strand cDNA synthesis kit (Applied Biosystems). Reverse transcription quantitative polymerase chain reaction was carried out with Standard SYBR Green qPCR Mix. The relative quantity was calculated by the 2-△△Ct method. The results for miRNA were normalized to U6 snRNA, and the results for DDIT4 were normalized to GAPDH (22). The primers are listed in Table 1 (14, 22).
| Name | Primer Sequence (5′-3′) |
|---|---|
| miR-642a-5p | F: GCGGTCCCTCTCCAAATGT; R: AGTGCAGGGTCCGAGGTATT |
| U6 | F: CTCGCTTCGGCAGCACA; R: AACGCTTCACGAATTTGCGT |
| DNA damage-inducible transcript 4 | F: CTTTGGGACCGCTTCTCGTC; R: GGTAAGCCGTGTCTTCCTCCG |
| GAPDH | F: CATGAGAAGTATGACAACAGCCT; R: AGTCCTTCCACGATACCAAAGT |
3.9. Western Blot
HepG2.2.15 cells were lysed with RIPA lysis buffer (Beyotime Institute of Biotechnology, China) containing 1% phenylmethanesulfonyl fluoride. Total protein, ranging from 20 - 50 mg, was separated on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel, transferred to a polyvinylidene fluoride membrane, and incubated with the primary antibody DDIT4 (1: 500, Abcam, Cambridge, MA, USA). Following incubation with horseradish peroxidase-labeled rabbit anti-mouse IgG (1: 2000, Abcam, ab6728), protein bands were visualized using an enhanced chemiluminescence system (Pierce Biotechnology Inc., Rockford, IL, USA) and imaged with a ChemiDoc XRS Plus Luminescence Image Analyzer (Bio-Rad). The density analysis of protein bands was performed using Image Lab software (Bio-Rad). GAPDH served as a control (23).
3.10. The Luciferase Method
The binding sites of miR-642a-5p and DDIT4 were predicted using the website https://cm.jefferson.edu/RNA22/Precomputed/. The 3’-untranslated region (3’-UTR) sequence of the DDIT4 gene was constructed, and the vectors were double-digested with SalI and BamHI. GenePharma (Shanghai, China) synthesized both wild-type (DDIT4-WT) and mutant (DDIT4-MUT) DDIT4-3′ UTRs containing the miR-642a-5p binding site. These sequences were amplified and cloned into a Luc promoter-containing pmirGLO luciferase reporter vector (Promega, Madison, WI, USA) to construct wild-type and mutant reporter vectors. Using cDNA from HepG2.2.15 cells as a template, 150 µg of luciferase reporter plasmid was co-transfected with miR-642a-5p mimic or mimic-NC (final concentration 50 nmol/L) using Lipofectamine® 2000 (Invitrogen) into HepG2.2.15 cells for 48 hours. The activities of firefly and Renilla luciferases were measured by a luciferase reporter assay (Promega Corporation, Madison, WI, USA). Firefly luciferase activity was normalized to Renilla activity. The ratio of firefly to Renilla luciferase activity in the same sample well (Firefly/Renilla luminescence) was used to evaluate the relative activity of luciferase. This assessment was repeated five times (24).
3.11. Statistical Analysis
Data analysis was conducted using SPSS 16.0 (IBM Corporation, New York, USA). Quantitative variables were presented as mean ± standard deviation (SD). Student's t-tests were utilized to evaluate differences between groups. The expression levels of miR-642a-5p and DDIT4 in LC tissue and adjacent non-cancerous liver tissue were compared using paired t-tests. The relationship between miR-642a-5p and DDIT4 expression was assessed using correlation analysis. One-way analysis of variance (ANOVA) was applied for comparisons among multiple groups. A P-value of less than 0.05 was considered to indicate statistical significance (25).
4. Results
4.1. miR-642a-5p is Downregulated and DNA Damage-Inducible Transcript 4 is Elevated in Hepatitis B Virus Liver Cancer Patients
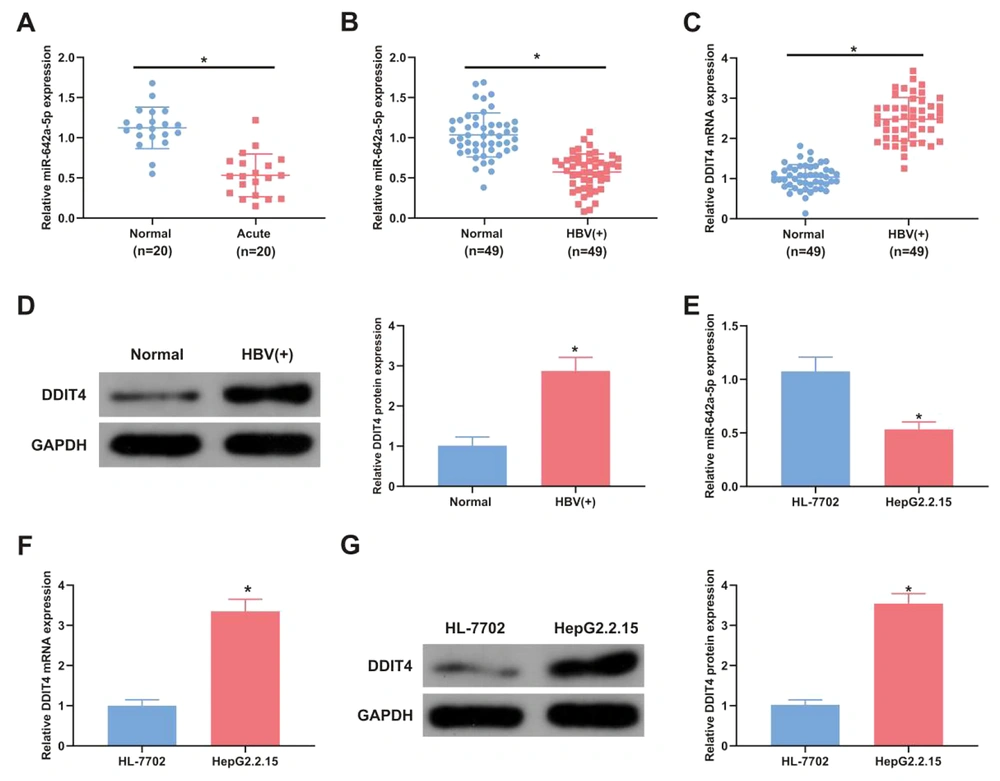
Serum miR-642a-5p levels were measured in 20 HBV-positive patients and 20 healthy controls to assess differences between these groups. The concentration of miR-642a-5p in the serum of HBV-positive patients was significantly lower than in healthy controls (P < 0.05) (Figure 1A). Additionally, miR-642a-5p and DDIT4 mRNA and protein levels were evaluated in the tissues of 49 patients with HBV-associated LC. In HBV-positive LC tissues, miR-642a-5p levels were decreased (P < 0.05), while DDIT4 mRNA and protein levels were increased compared with those in the healthy control group (P < 0.05) (Figure 1B - D).
miR-642a-5p is silenced in hepatitis B virus (HBV) liver cancer (LC) patients, while DNA damage-inducible transcript 4 (DDIT4) is in the opposite. A, the relative expression of miR-642a-5p in the serum of healthy individuals (control, n = 20) and HBV acute infection patients (acute infection, n = 20); B, miR-642a-5p in tissues of healthy individuals and HBV-positive patients (n = 49 per group); C, DDIT4 mRNA in the tissues of healthy individuals and HBV-positive patients; D, DDIT4 protein in the tissues of healthy individuals and HBV-positive patients; E, miR-642a-5p in HL-7702 and HepG2.2.15 cells; F, DDIT4 mRNA in HL-7702 and HepG2.2.15 cells; G, DDIT4 protein expression in HL-7702 and HepG2.2.15 cells. All experiments at the cellular level were repeated five times. * P < 0.05.
The association between miR-642a-5p expression and clinicopathological characteristics of HBV-positive HCC patients was analyzed. Patients with HBV-positive HCC, confirmed by positive HBV tests and pathological biopsies, exhibited significantly lower miR-642a-5p expression in stages III and IV and in tumors larger than 5 cm compared to those in stages I and II and tumors smaller than 5 cm (Table 2).
| Variables | Number of Patients (n = 49) | miR-642a-5p | P-Value | |
|---|---|---|---|---|
| The Declined (n = 24) | The Elevated (n = 25) | |||
| Gender | 0.5512 | |||
| Female | 32 (57) | 17 | 15 | |
| Male | 17 (43) | 7 | 10 | |
| Age | 0.2516 | |||
| 45 or more | 21 (67) | 8 | 13 | |
| Less than 45 | 28 (37) | 16 | 12 | |
| TNM stage | 0.0003 a | |||
| I + II | 19 (39) | 3 | 16 | |
| II + VI | 30 (61) | 21 | 9 | |
| Tumor size | 0.0001 a | |||
| 5 cm or more | 23 (47) | 19 | 4 | |
| Less than 5 cm | 26 (53) | 5 | 21 | |
a P ≤ 0.001 was considered statistically significant.
miR-642a-5p and DDIT4 levels were also measured in the human normal liver cell line HL-7702 and the HBV-positive HepG2.2.15 cell line. Compared with HL-7702 cells, the levels of miR-642a-5p in HepG2.2.15 cells were decreased (P < 0.05) (Figure 1E), and DDIT4 mRNA and protein levels were increased (P < 0.05) (Figure 1F and G).
4.2. miR-642a-5p Represses Hepatitis B Virus DNA Replication
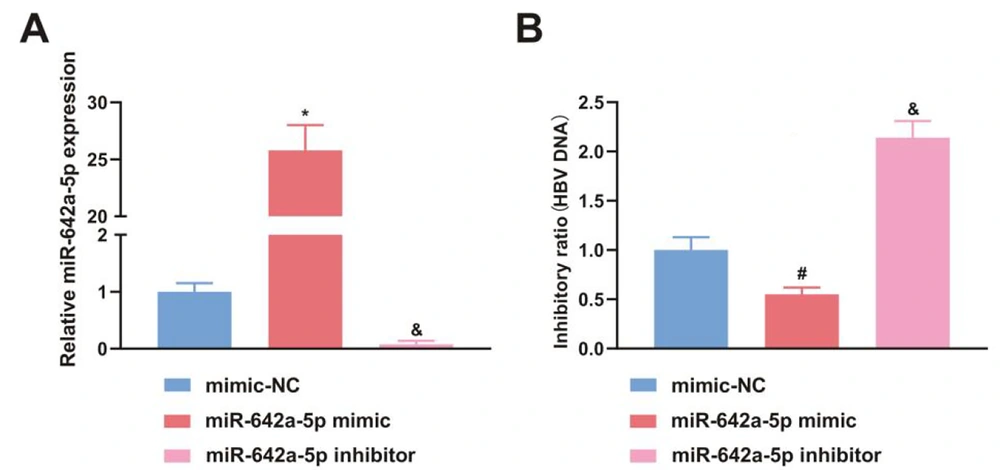
miR-642a-5p mimic was transfected into HBV-infected HepG2.2.15 cells to investigate the effect of miR-642a-5p on HBV DNA replication. In cells transfected with the miR-642a-5p mimic, miR-642a-5p expression was significantly increased (P < 0.05) (Figure 2A), while HBV DNA replication levels were decreased compared to those in cells transfected with miR-NC and the miR-642a-5p inhibitor (P < 0.05) (Figure 2B).
miR-642a-5p upregulation suppresses hepatitis B virus (HBV) DNA replication. A, miR-642a-5p after transfection of mimic-NC, miR-642a-5p mimic and miR-642a-5p inhibitor in HepG2.2.15 cells; B, HBV DNA copy number of HepG2.2.15 cells after transfection with mimic-NC, miR-642a-5p mimic and miR-642a-5p inhibitor. All experiments at the cellular level were repeated five times. *, # P < 0.05 versus the miR-642a-5p mimic and the mimic-NC, & P < 0.05 versus the miR-642a-5p inhibitor and mimic-NC.
4.3. miR-642a-5p Upregulation or DNA Damage-Inducible Transcript 4 Downregulation Restrains Hepatitis B Virus Hepatocellular Carcinoma Proliferation
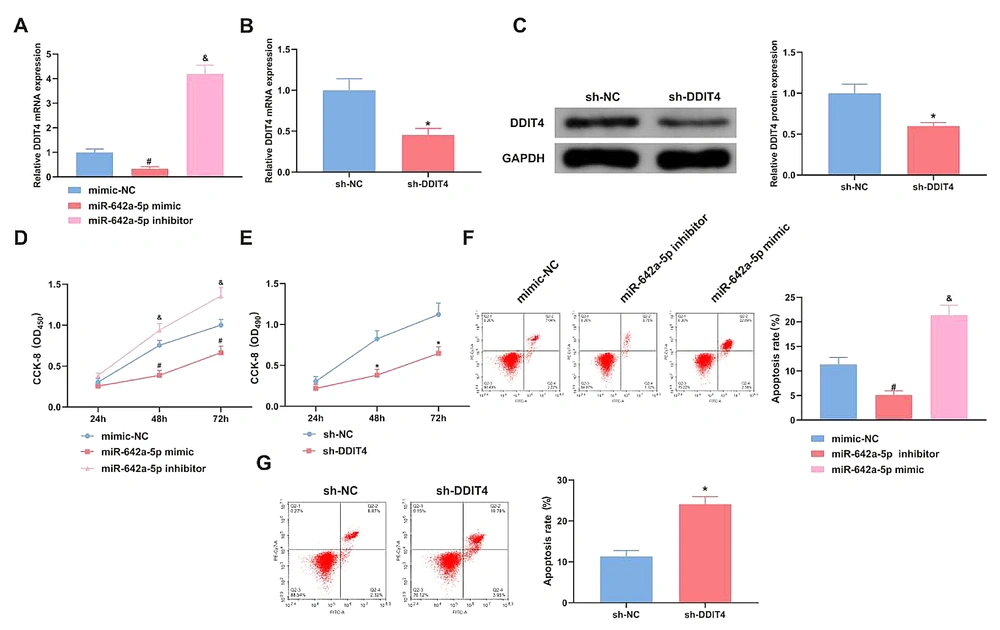
miR-642a-5p mimic/inhibitor and DDIT4 plasmids were transfected to regulate miR-642a-5p and DDIT4. After transfection with miR-642a-5p mimic, miR-642a-5p levels were elevated in HepG2.2.15 HCC, which significantly reduced the mRNA level of DDIT4 in these cells. The results showed that the miR-642a-5p mimic significantly inhibited DDIT4 expression (P < 0.05) (Figure 3A). In cells transfected with the si-DDIT4 plasmid, both DDIT4 mRNA and protein levels were reduced (P < 0.05) (Figure 3B and C). Cell proliferation assays demonstrated that the proliferation rate of HepG2.2.15 HCC was significantly decreased following transfection with either miR-642a-5p mimic or si-DDIT4 (P < 0.05) (Figure 3D and E). Furthermore, either increasing miR-642a-5p or silencing DDIT4 promoted apoptosis in HepG2.2.15 HCC (P < 0.05) (Figure 3F and G). In conclusion, elevating miR-642a-5p or silencing DDIT4 inhibited HBV proliferation in HCC.
Elevation of miR-642a-5p or silence of DNA damage-inducible transcript 4 (DDIT4) suppresses the proliferation of hepatitis B virus (HBV) hepatocellular carcinoma (HCC). A, effect of miR-642a-5p on DDIT4 expression; B, DDIT4 mRNA in HepG2.2.15 cells transfected with sh-NC and sh-DDIT4; C, DDIT4 expression after sh-DDIT4 transfection; D - E, HepG2.2.15 cell proliferation detected by CCK-8 assay; F - G, HepG2.2.15 cell apoptosis analyzed by flow cytometry. All experiments at the cellular level were repeated five times. * P < 0.05, # versus the miR-642a-5p mimic and the mimic-NC, & P < 0.05 versus the miR-642a-5p inhibitor and the mimic-NC.
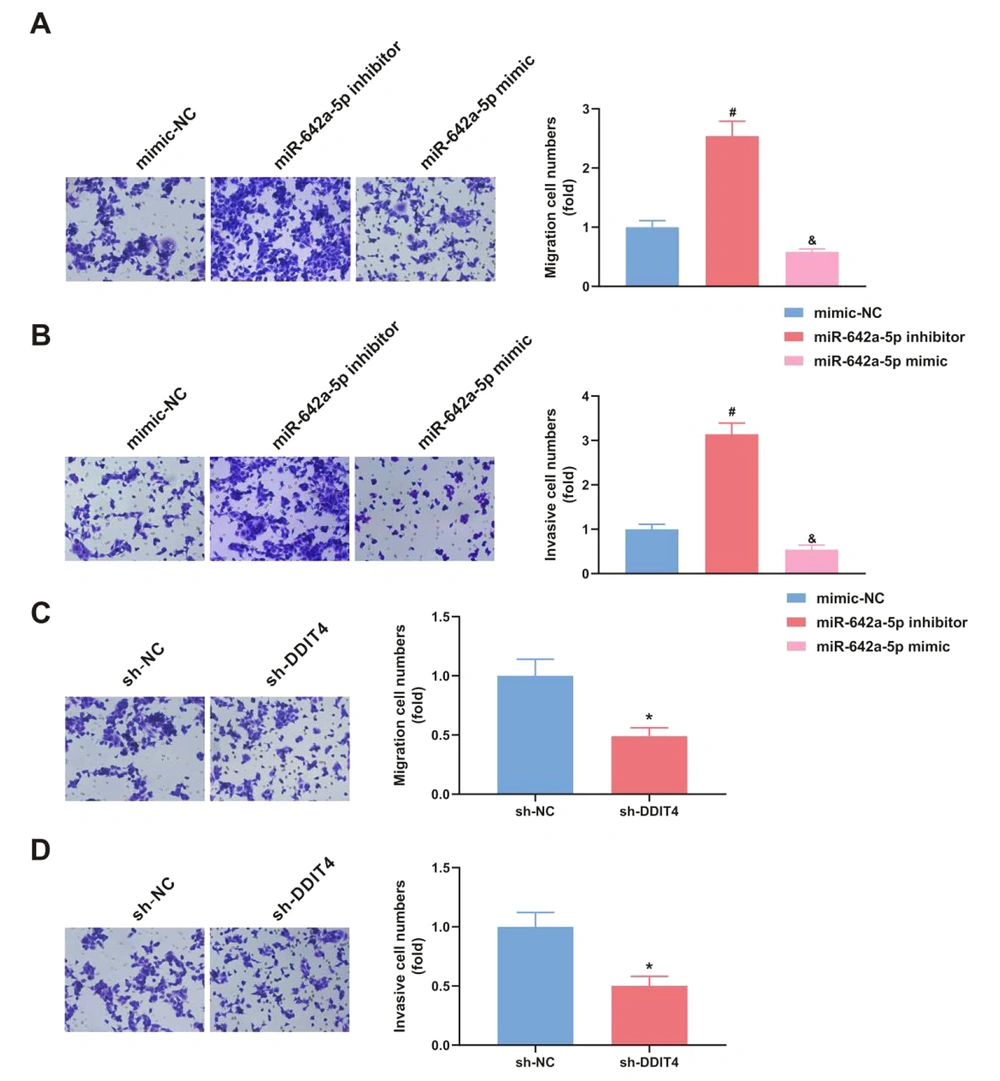
4.4. miR-642a-5p Upregulation or DNA Damage-Inducible Transcript 4 Downreuglation Represses Hepatitis B Virus Hepatocellular Carcinoma Progression
The progression of HCC was decreased following the transfection of miR-642a-5p mimic (P < 0.05), whereas the transfection of miR-642a-5p inhibitor had the opposite effect (P < 0.05) (Figure 4A and B). Additionally, si-DDIT4 suppressed HBV HCC progression (P < 0.05) (Figure 4C and D).
Upregulation of miR-642a-5p or silence of DNA damage-inducible transcript 4 (DDIT4) represses hepatitis B virus (HBV) hepatocellular carcinoma (HCC) migration and invasion. A-B, elevation of miR-642a-5p restrained HCC migration and invasion; C-D, silence of DDIT4 repressed HCC migration and invasion. All experiments at the cellular level were repeated five times. * P < 0.05, # versus the miR-642a-5p mimic and the mimic-NC, & P < 0.05 versus the miR-642a-5p inhibitor and the mimic-NC.
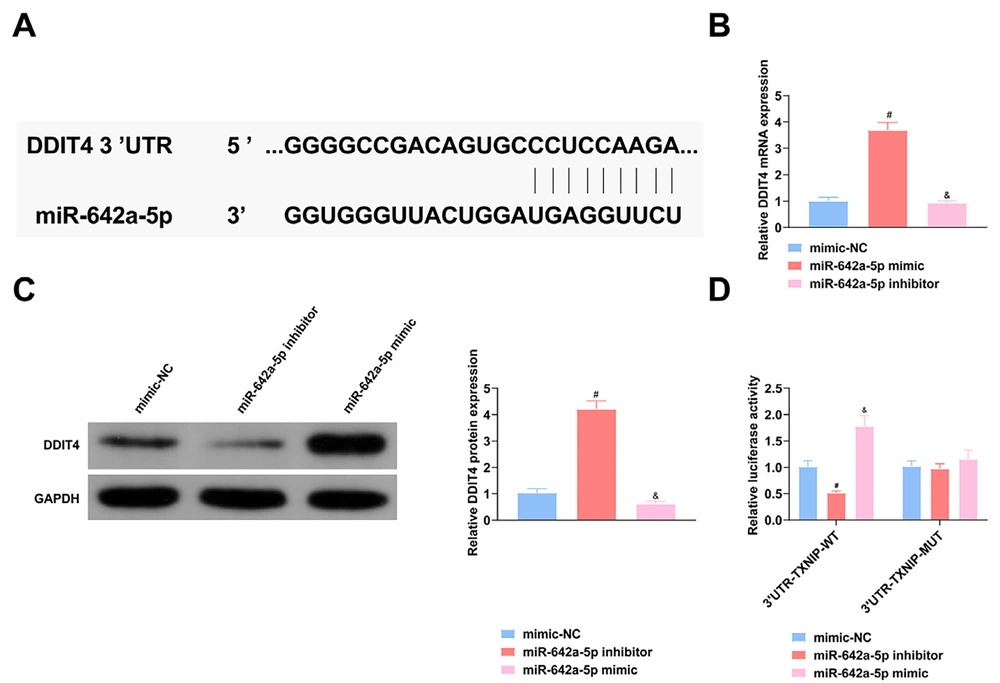
4.5. DNA Damage-Inducible Transcript 4 Is the Target of miR-642a-5p
The miR-642a-5p mimic inhibited DDIT4 mRNA and protein expression, while the miR-642a-5p inhibitor had the opposite effect (P < 0.05) (Figure 5A), demonstrating that miR-642a-5p regulates DDIT4 at both mRNA and protein levels. Binding sites of miR-642a-5p and DDIT4 were identified on the website https://cm.jefferson.edu/RNA22/Precomputed/ (Figure 5B). DNA Damage-inducible transcript 4 was confirmed as a direct target of miR-642a-5p. Additionally, the miR-642a-5p mimic suppressed the luciferase activity of the wild-type reporter (Figure 5C), further confirming that DDIT4 is the target gene of miR-642a-5p.
miR-642a-5p targets DNA damage-inducible transcript 4 (DDIT4). A, Schematic diagram of the 3’UTR of DDIT4 and the putative target of miR-642a-5p; B, DDIT4 mRNA in hepatocellular carcinoma (HCC) transfected with miR-642a-5p mimic; C, DDIT4 protein in HCC transfected with miR-642a-5p mimic; D, luciferase activity of HepG2.2.15 cells. All experiments at the cellular level were repeated five times. * P < 0.05, # versus the miR-642a-5p mimic and the mimic-NC, & P < 0.05 versus the miR-642a-5p inhibitor and the mimic-NC.
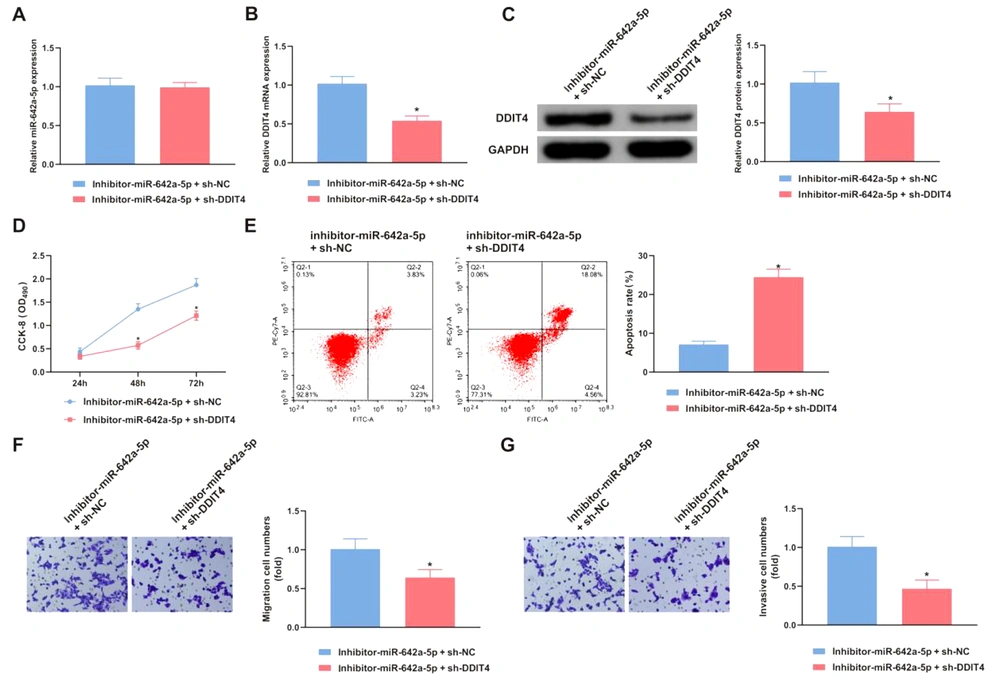
4.6. Silence of DNA Damage-Inducible Transcript 4 Turns Around the Function of miR-642a-5p on Hepatitis B Virus Hepatocellular Carcinoma
mRNA and protein levels of miR-642a-5p and DDIT4 were measured in HCC cells transfected with either inhibitor-miR-642a-5p + sh-NC or inhibitor-miR-642a-5p + sh-DDIT4. The cells treated with inhibitor-miR-642a-5p + sh-DDIT4 exhibited lower DDIT4 expression compared to those treated with inhibitor-miR-642a-5p + sh-NC (Figure 6A, B and C). Furthermore, functional assay results revealed that downregulating DDIT4 mitigated the effects of miR-642a-5p silencing on the proliferation, apoptosis, invasion, and migration of HepG2.2.15 cells (P < 0.05) (Figure 6D - G).
DNA damage-inducible transcript 4 (DDIT4) turns around the action of miR-642a-5p. A, miR-642a-5p in HepG2.2.15 cells transfected with inhibitor-miR-642a-5p + sh-NC and inhibitor-miR-642a-5p + sh-DDIT4; B, DDIT4 mRNA in HepG2.2.15 cells transfected with inhibitor-miR-642a-5p + sh-NC and inhibitor-miR-642a-5p + sh-DDIT4; C, DDIT4 protein in HepG2.2.15 cells transfected with inhibitor-miR-642a-5p + sh-NC and inhibitor-miR-642a-5p + sh-DDIT4; D, proliferation of HepG2.2.15 cells transfected with inhibitor-miR-642a-5p + sh-NC and inhibitor-miR-642a-5p + sh-DDIT4; E, apoptosis of HepG2.2.15 cells transfected with inhibitor-miR-642a-5p + sh-NC and inhibitor-miR-642a-5p + sh-DDIT4; F, migration of HepG2.2.15 cells transfected with inhibitor-miR-642a-5p + sh-NC and inhibitor-miR-642a-5p + sh-DDIT4; G. invasion of HepG2.2.15 cells transfected with inhibitor-miR-642a-5p + sh-NC and inhibitor-miR-642a-5p + sh-DDIT4. All experiments at the cellular level were repeated five times. * P < 0.05.
5. Discussion
Hepatitis B virus infection is recognized globally as a major cause of HCC, with 0.4% to 0.6% of chronically infected HBV patients being diagnosed with HCC annually, a rate significantly higher than in non-infected individuals. Understanding the pathogenesis of HBV-associated HCC and the roles and signaling pathways of miRNAs in HCC is crucial. miRNAs are closely associated with the development of HBV-related HCC, exhibiting varying expression levels and diverse mechanisms of action. Therefore, elucidating the mechanisms of HBV-induced HCC is critical in identifying new therapeutic targets. This study highlights the inhibitory role of miR-642a-5p in the proliferation, migration, and HBV DNA replication of HBV-associated HCC by targeting DDIT4.
Researchers have demonstrated that miRNAs, due to their high stability in blood, can serve as non-invasive biomarkers for cancer detection. For instance, miR-375 has been identified as a serological marker for HCC. Furthermore, numerous studies have reported miRNA dysregulation in HBV-infected HCC, including miR-106b (26), miR-21 (27), and miR-221 (28). Aberrant miRNA expression profiles in HCC can potentially serve as biomarkers for diagnosing HCC, as well as predicting its metastasis and invasion. Contrary to previous studies, miR-642a-5p functions as a tumor suppressor in HBV-associated HCC. It is downregulated in the serum, tissues, and cells of patients with HBV HCC. An analysis of the relationship between miR-642a-5p and the clinicopathological features of HBV-positive HCC patients showed that miR-642a-5p expression in patients with TNM stages III and IV and tumor size greater than 5 cm was significantly lower than in those with stages I and II and tumor size less than 5 cm. This indicates that miR-642a-5p is expressed at low levels in patients with HBV-associated HCC and could serve as a crucial screening indicator for cancer progression.
In this study, we selected para-cancer normal tissues, also known as non-tumor tissues, as controls. These are commonly used in tumor-related studies because they are in a preneoplastic state that consists of morphologically normal but molecularly altered cells prone to accumulating oncogenic events. We primarily analyzed clinical samples for oncogene testing. Using para-cancer normal tissue as a control has several advantages. It reflects the microenvironment surrounding the tumor, which may aid in developing clinical strategies and specific interventions for preventing recurrence. Moreover, comparing samples from the same individual may minimize individual-specific and anatomical site-specific effects.
In conclusion, our findings support that miR-642a-5p has a tumor-suppressive function in the development of HCC. Hepatitis B virus, a hepatotropic DNA virus, is a major cause of HCC. The role of miRNAs in regulating host-virus interactions involved in HBV infection, replication, and associated diseases is increasingly recognized. For instance, miR-152 has been shown to cause hypermethylation of HBV DNA in HCC cell lines (29). In our study, increased levels of miR-642a-5p suppressed HBV DNA copy numbers in HBV-infected HepG2.2.15 cells. Up-regulation of miR-642a-5p also led to reduced proliferation, migration distance, and fewer cells penetrating the membrane in HCC cell assays. These findings suggest that miR-642a-5p plays a significant tumor-suppressive role in HBV-associated HCC.
miR-642a-5p is found to be increased in the serum of patients with acute respiratory distress syndrome (30) and pterygium (31). Additionally, it is upregulated in renal cell carcinoma (32), whereas it is downregulated in cervical cancer cells (33). Dysregulation of miR-642a-5p has also been observed in human prostate cancer (34) and pancreatic cancer (35). While the expression profile and biological function of miR-642a-5p have been elucidated in some human cancers, its biological role in tumors remains underexplored, particularly its mechanisms in HBV-infected HCC. Tumorigenesis often involves disruptions in the processes of cell proliferation and apoptosis. miRNAs inhibit their target genes by binding to the 3'UTR sequences through the miRISC complex, promoting mRNA degradation or inhibiting its translation (36).
In this study, bioinformatics analysis indicated that miR-642a-5p could bind to the 3'UTR of DDIT4 mRNA and inhibit its expression. Dual-luciferase assay results confirmed a targeted regulatory relationship between miR-642a-5p and DDIT4 in HepG2.2.15 cells. Kunadirek et al. (17) identified DDIT4 expression as a potential diagnostic biomarker for distinguishing HCC from non-HCC conditions through a microarray dataset, highlighting its specificity for HCC. Our findings show that upregulation of DDIT4 reverses the inhibitory effects of miR-642a-5p on HBV DNA replication and cell progression. This not only substantiates the targeting relationship between miR-642a-5p and DDIT4 but also opens new avenues for targeted therapy and clinical diagnosis in HCC.
At the same time, increased DDIT4 has been significantly associated with poor prognosis in acute myeloid leukemia, glioblastoma multiforme, breast cancer, lung cancer, and skin cancer. However, elevated DDIT4 is linked with improved prognosis in gastric cancer but not in ovarian cancer (37). These findings suggest that DDIT4 functions differently across various cancer types. In this study, DDIT4 was elevated in HBV-stimulated HCC, and it was considered a novel biomarker for HBV HCC. However, research on the potential role of DDIT4 in HCC, especially HBV-infected HCC, remains limited. DDIT4 negatively regulates the mammalian target of rapamycin (mTOR) signaling pathway, which plays a crucial role in tumor invasion by controlling various functional and biological processes, including cell propulsion. Thus, inhibiting the mTOR pathway is an effective cancer treatment strategy (38). In the AKT/mTORC signaling pathway, DDIT4 influences TSC2 phosphorylation through protein phosphatase 2A, thereby inhibiting mTORC and impacting cancer treatment. Infection analysis indicated that DDIT4 levels in HBV-positive HCC patients were significantly higher compared to HBV-negative HCC patients and healthy individuals, suggesting its potential utility in distinguishing early HBV-positive HCC from non-HBV-positive HCC.
Therefore, miR-642a-5p and DDIT4 may become new biological targets for the early diagnostic or therapeutic targeting of HBV HCC in the future, providing reference value for basic research and clinical personalized treatment of HCC. However, the current study acknowledges deficiencies in the investigation of HCC, such as the lack of strong diagnostic specificity, and the non-specific expression of miR-642a-5p and DDIT4 in other tumors, including pancreatic and skin cancers. Additionally, the mTOR pathway will be the focus of future investigations of the miR-642a-5p/DDIT4 axis in HBV HCC. This study is still in the preliminary stage with a small sample size, and the potential mechanism of the miR-642a-5p/DDIT4 axis in HBV HCC remains unclear. The interaction between miR-642a-5p and its target gene DDIT4 needs to be verified by further in vivo and in vitro experiments. Therefore, more in vivo experiments and clinical studies are needed to further confirm the clinical application of the miR-642a-5p/DDIT4 axis in HBV HCC, and to provide a theoretical basis for early screening, diagnosis, treatment, and prognostic evaluation of HCC.
5.1. Conclusions
In summary, miR-642a-5p is silenced in HBV-positive HCC tissues, and it targets DDIT4 to inhibit both HBV DNA replication and the progression of HBV HCC. Additionally, this study introduces a new miRNA that could serve as a novel biomarker for the clinical diagnosis and targeted therapy of LC, aiding in understanding the mechanism of HBV HCC development.