1. Background
Endometriosis is a common female disorder characterized by endometrial-like tissue lesions outside the uterus, affecting 10 - 15% of women of reproductive age and 20 - 50% of infertile women (1, 2). The frequency of this disorder among infertile Iranian women has been reported as 29% (3). Endometriosis appears during reproductive years and is associated with a wide range of symptoms (1). Despite extensive research, the pathogenesis and molecular basis of this disease remain unknown. Identifying a single factor that explains the pathogenesis of this disease is challenging [1]. It is believed that genetic and immunological factors (4), hormonal factors, and inflammation are involved in the regulation of endometriosis (5). Evidence suggests a complex two-way interaction between endometriosis and the microbiome (6).
The effect of microbiota on the epigenetic, immunological, or biochemical functions of the host has also been debated (4). Importantly, antibiotics and probiotics have been found effective in the treatment of endometriosis, indicating a connection between the microbiome and this disease (7). Recent studies have demonstrated changes in the microbiota of the genital system with the progression of endometriosis and the alleviation of disease symptoms with antibiotic therapy (7). Lactobacillus is the dominant bacterium in the female reproductive system. By modulating inflammatory processes and producing different metabolites such as lactic acid, H2O2, and bacteriocin, it inhibits the colonization of pathogenic bacteria (8). Endometriosis progresses by reducing the number of lactobacilli and increasing the number of other bacteria (7).
2. Objectives
This study aims to examine the bacterial populations in the cervix and endometrium of women with endometriosis compared to women without the disease using culture-based methods and to compare the frequency of lactobacilli in patients with and without endometriosis using quantitative real-time PCR (qPCR) assay. By reliably quantifying bacteria, qPCR may improve our understanding of the association between Lactobacillus and endometriosis. Since there is a lack of understanding of the relationship between them, identifying this relationship can help elucidate the pathogenesis and ultimately lead to the development of non-invasive testing and diagnosis. Additionally, it serves as a fundamental step toward developing new treatment strategies.
3. Methods
3.1. Sampling
One hundred women aged 18 to 40 who were referred to the IVF Department of Arash Women's General Hospital in Tehran from July 2021 to July 2022 were divided into two groups: Fifty women with ultrasound-confirmed endometriosis who were seeking treatment for infertility, and 50 healthy women in the control group who were referred for egg donation. Exclusion criteria included infection of the genital tract in the last 3 months, use of hormonal contraception or IUD, use of antibiotics or probiotics in the last 8 weeks, abnormal pap smear results in the last 3 years, vaginal bleeding, use of vaginal drugs in the last 3 weeks, and sexual activity in the last week. The treatment protocol for both groups involved gonadotropin-releasing hormone (GnRH) agonists.
3.2. Sample Collection
The samples were collected on the 3rd to 7th day after menstruation. The endometrial samples were collected by a gynecologist using a pipelle (Medbar) under strict aseptic conditions. For cervical sampling, during a speculum examination, two separate dacron swabs were gently rotated in the cervix for five circles to obtain secretions.
3.3. DNA Extraction
DNA was extracted from endometrial specimens using the GeneAll kit as instructed by the supplier (GeneAll Biotechnology, Korea). The cervical specimens were centrifuged at 3500 × g for 5 minutes at 4°C, and then the sediments were used for DNA extraction using the same kit. DNA samples were stored at -20°C.
3.4. Quantitative Real time Polymerase Chain Reaction
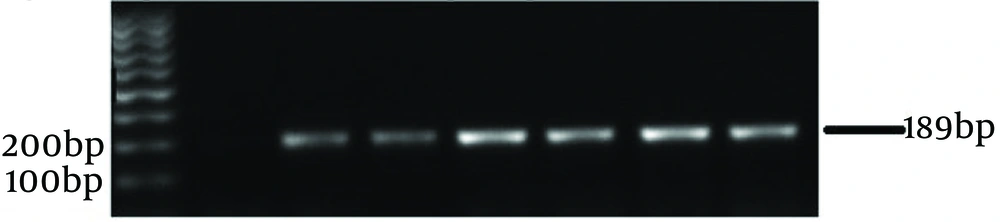
The thermocycling conditions consisted of an initial melting step at 95°C for 20 seconds, followed by 55°C for 20 seconds, and 72°C for 20 seconds, in a total volume of 20 μL. This volume included 3 μL DNA sample, 0.8 μL of each primer (9) (0.4 μmol/L), 10 μL SYBR Green PCR master mix (2×), and 5.6 μL double-distilled water. The experimental data were performed at least in triplicate, and results were expressed as mean ± SEM. To prepare an external standard, the DNA was serially diluted in double-distilled water ranging from 106 to 103 copies according to ABI guidelines on "Creating Standard Curves with Genomic DNA Templates for Use in Quantitative PCR." Aliquots of each dilution were stored at -20°C until use. A non-template control was used in each qPCR experiment as a negative control.
3.5. Polymerase Chain Reaction for Detection of 16S rRNA Genes
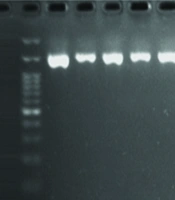
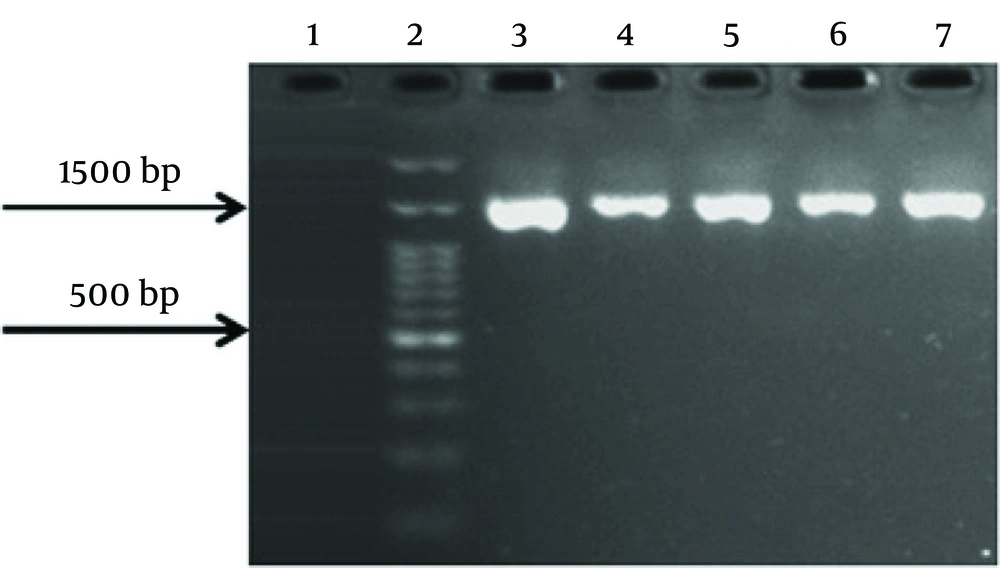
The samples that were negative for 16S Lactobacillus were checked with two primers (10, 11) listed in Table 1 to ensure the correctness of the extraction. The PCR amplification mixture was 25 μL, with an initial denaturation step at 94°C for 5 minutes, followed by 40 cycles at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1.5 minutes, and a final extension step at 72°C for 10 minutes. Each analysis included bacterial positive and negative controls.
3.6. Isolation and Identification of Microorganisms
The samples were cultured in two ways. The first group was cultured immediately after sampling, and the second group was incubated in Thioglycollate Broth medium (QUELAB company, Canada) for 48 hours at 37ºC and then cultured on the appropriate medium. The samples were first examined in nutrient media such as blood agar and chocolate agar. Colonies grown on these media were then recultured on several different selective and specific media, such as eosin methylene blue (EMB) and mannitol salt agar, and incubated for 24 to 72 hours at 37ºC. Confirmatory methods including Gram staining and phenotypic tests such as catalase, urease, growth on Simon citrate, TSIA and SIM media, production of oxidase, DNase, lysine decarboxylase, and methyl red (MR)/Voges-Proskauer were used for identification. Rogosa and Sharpe (MRS) and brain-heart infusion (BHI) agar were used to isolate Lactobacillus, with 0.05% L-cysteine added to the MRS and BHI media. The plates were incubated for 24 hours at 37ºC in anaerobic and microaerophilic conditions (12). The Vitek system was also used to identify the samples, and one sample was sent to Genomin Iran for sequencing.
3.7. Statistical Analysis
Data were analyzed using SPSS software, version 26 (SPSS, Chicago, IL, USA). Differences between the healthy controls and patients were compared using the chi-square test. Data were visualized with GraphPad Prism version 9 (GraphPad Software, Inc., San Diego, CA, USA).
4. Results
Overall, 100 women were examined, 50 of whom had endometriosis confirmed by ultrasound, while 50 healthy women served as the control group. In this study, a total of 200 samples were examined, including 100 cervical swab samples and 100 endometrial biopsy samples. The samples that were immediately cultured without enrichment did not grow. The results of the samples enriched in Thioglycolate broth for 48 hours are listed in Table 2. The bacteria found in the cervix and endometrium were very similar. A total of 17 species of bacteria and 3 yeasts were isolated. Fifty-eight and 49 isolates were obtained from the cervix and endometrium of individuals with endometriosis, respectively, while 34 and 23 isolates were obtained from the control group. In the case group, no isolates were found in 4 individuals, while in the control group, no isolates were found in 23 individuals. Three different types of bacteria were isolated from 1 individual in the case group, while in the control group, 5 individuals had three different types of bacteria isolated. The chi-square test revealed a significant relationship between the frequency of enterococci and Enterobacteriaceae in the cervix and endometrium and endometriosis (P < 0.05).
| Bacteria | Case Group | Control Group | ||
|---|---|---|---|---|
| Cervix | Endometrial | Cervix | Endometrial | |
| Mycoplasma hominis ATCC23114 | - | - | 1 | - |
| Lactobacillus | 4 | 4 | 6 | 6 |
| Enterococcus | 15 | 16 | 6 | 5 |
| Escherichia coli | 12 | 12 | 3 | 3 |
| Klebsiella pneumonia | 6 | 6 | 1 | - |
| Edwardsiella tarda | 1 | - | - | - |
| Proteus mirabilis | 1 | - | - | - |
| Citrobacter | - | - | - | 1 |
| Staphylococcus aureus | 3 | - | - | - |
| S. epidermidis | 2 | 1 | 2 | - |
| S. saprophyticus | 1 | 2 | 2 | 1 |
| Staphylococcus spp. | 3 | 4 | 5 | 3 |
| Non-hemolytic Streptococcus | 4 | 4 | 5 | 3 |
| Streptococcus agalactiae | 2 | - | 1 | 1 |
| S. anginous | 1 | - | - | - |
| S. pneumoniae | 2 | - | 2 | - |
| Sphingomonas paucimobilis | 1 | - | - | - |
Quantitative PCR was performed to check the number of lactobacilli in the endometrium and cervix. Each qPCR test was repeated three times. The average results were used to create graphs and compare the data. Based on specific qPCR of endometrial Lactobacillus species, 33 (66%) of healthy women and 24 (48%) of women with endometriosis were positive. Fifty-seven percent of endometrial samples were positive for Lactobacillus. The negative samples were re-checked with two sets of primers for 16S rRNA genes (shown in Figures 1 and 2). The results showed that specimens from 5 individuals (3 controls and 2 cases) were negative for bacterial 16S rRNA genes. It should be noted that these individuals also had negative culture results. The number of Lactobacillus bacteria in cervical samples was measured using the qPCR method, and samples with fewer than 10 bacteria were considered negative.
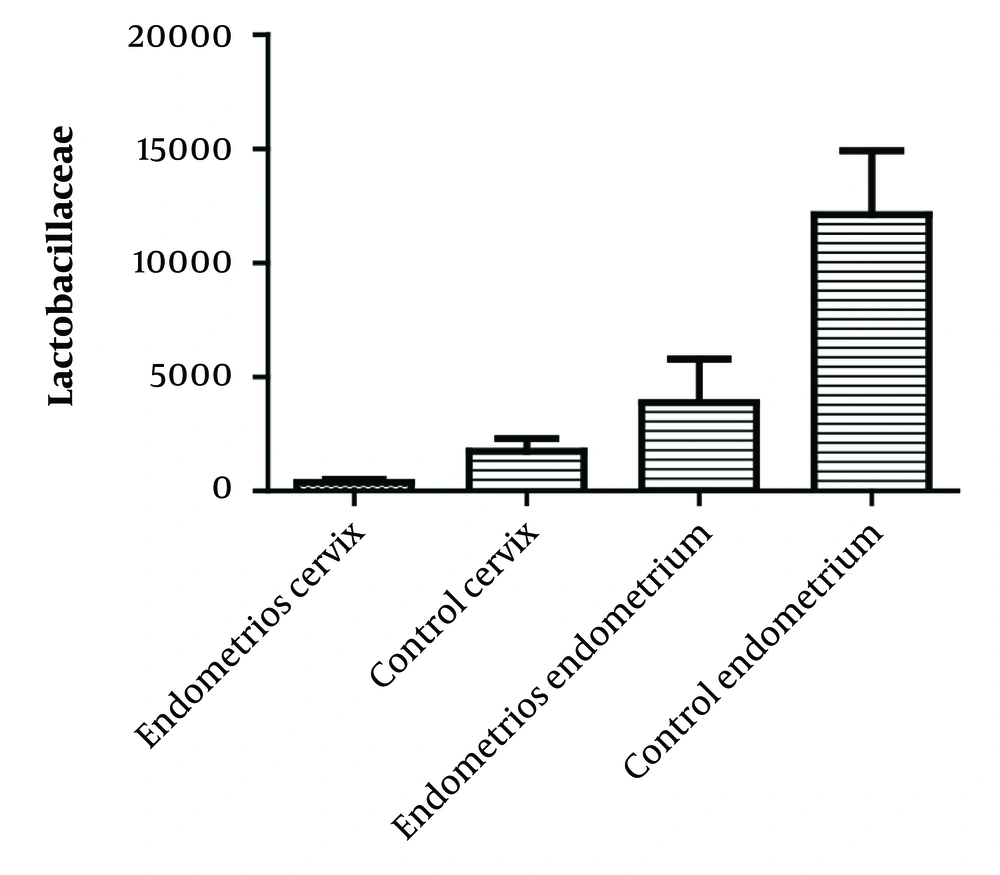
Ninety percent (48 individuals) of the control group and 70% (44 individuals) of the case group had qPCR results above 10. The average number of lactobacilli in the cervical samples of the case and control groups were 377 and 1734, respectively. In the endometrial samples, the average number was 3875 in the affected group and 12108 in the control group. The average number of lactobacilli in cervical and endometrial samples is shown in Figure 3. A significant relationship (P < 0.05) was found between the colonization of the uterus and cervix with Enterococcus spp. and Enterobacteriaceae and the reduction of lactobacilli in the case group.
5. Discussion
Endometriosis is a chronic inflammatory disease characterized by the presence of endometrial tissue in other organs than uterus. It results in pelvic pain and infertility and deteriorate the quality of life (5, 13). Despite the high number of patients, diagnosis is usually delayed for years, misdiagnosis is common, and effective treatment takes time to provide. It is necessary to investigate the factors triggering the disease in particular the role of microbiota in relation to disease symptoms (13, 14). In this study, we compared the microbiota of the endometrium from endometrial biopsy and cervical swap samples collected from patients with endometriosis and control group.
Using qPCR, we demonstrated the abundance of lactobacilli in cervix and endometrium in control group, as well as the relation between the colonization of Enterococcus spp. and Enterobacteriaceae in the uterus and cervix and reduction of lactobacilli in case group. It is also important to mention that 5 endometrial biopsies (2 patients and 3 control groups) did not yield PCR products for the 16S rRNA gene. This indicates that some people may not have microbiota in endometrium. In a similar study conducted by Wessels et al., three biopsy samples were negative for the 16S rRNA gene in PCR assay (15). Disease is consistently associated with reduction of lactobacilli and increase in bacteria involved in vaginosis and other opportunistic infections (7, 16). Lactobacilli produce various substances that prevent the growth of pathogenic bacteria. Inhibitors include lactic acid, bacteriocins and hydrogen peroxide (8, 17).
In the present study, the frequency of lactobacilli detected by culture method in the case group was 8%, which is significant. However, the frequency of lactobacilli detected by real-time quantitative PCR method in this group were 40% (from endometrium) and 70% (cervix samples). There was a significant relationship between the frequency of Lactobacillus and endometriosis as determined by real-time quantitative PCR method. The reason for higher sensitivity of PCR over the culture is its ability in detection of the nucleic acids, regardless of viability of bacterial cells. Similar finding has also been reported (18).
The composition of microbiome detected by culture from the cervix and endometrium was similar in our study confirming the results of Chen et al. and Winters et al. (19, 20). Samples that were directly cultured without enrichment did not grow, possibly due to the lower number of bacteria present in the uterus and upper endocervix (10,000-fold) comparing vagina. The reasons for difference was either the role of cervix as it functions as a filter or clears ascending bacteria by the endometrial immune response, or a combination of both (17, 21).
There was a significant relationship between the number bacteria belonging to the Enterobacteriaceae family and Enterococcus spp. in the endometrium and cervix and occurrence of endometriosis, which is consistent with previous studies. Khan et al., reported a relationship between intrauterine microbial colonization of endometrial samples and endometriosis comparing with control group. In their study the number of Enterococcus spp. and E. coli CFUs in the endometrial samples of women with endometriosis was significantly higher than control (5).
Using NGS analysis on cervical mucus from women with and without endometriosis, Akyama et al., reported that Enterobacteriaceae family members were found in significant amounts in women with endometriosis (22). In another study Cojocaru proved that endometriosis is associated with increased presence of members of Enterobacteriaceae family (4). Similarly, the incidence of endometriosis was related to the increase in the presence of Enterobacteriaceae, particularly E. coli, in different parts of the genital tract (6). Using PCR on specimens from the deep lesions of endometrium, up to 50 percent of patients with endometriosis were positive for Enterococcus spp. (16). In Khan's study in 2016, Enterobacteriaceae and Streptococcaceae were documented as the most important organisms in endometriosis group by real-time PCR (23). Increase in number of Enterococcus spp., and E. coli in women with endometriosis was also reported by other investigators (24-26).
5.1. Conclusions
The reduction of lactobacilli and the increase of other bacteria in people with endometriosis confirm studies aiming to transform dysbiosis into a favorable genital microenvironment using Lactobacillus and other probiotics. This approach is potentially effective for the prevention and even treatment of those suffering from this disease. Furthermore, the findings of this study provide a basis for further research to investigate the intrauterine colonization of different bacteria and their role in the occurrence of endometriosis. Such research could lead to the development of non-invasive diagnostic and treatment options.