1. Background
The viable but non-culturable (VBNC) state is a microbial survival strategy in response to unfavorable physicochemical conditions, characterized by the loss of culturability of microbial cells in standard culture media while maintaining their metabolic activity (1). Bacterial cells are capable of entering the VBNC state in response to various physicochemical stresses, including starvation (2), incubation outside the natural growth temperature range (3-5), elevated and lowered osmotic concentrations (6, 7), mineral salts (8), exposure to UV radiation (9), heavy metals (10), food preservatives (11, 12), etc. Unlike laboratory conditions, bacteria in nature spend most of their lives starved or nutrient-limited. Over the past two decades, microbial ecologists have stated that less than 1% of all bacteria present in nature can grow on conventional bacterial culture media. Some believe that one reason for this is the entry of bacteria into the VBNC state (13).
Shigella flexneri is highly host-adapted and only infects humans and some other mammals. Shigella flexneri is the main agent of shigellosis (bacillary dysentery), a clinical syndrome caused by the invasion of Shigella species into the epithelial lining of the distal ileum, colon, and rectum (14). The World Health Organization's analysis of foodborne diseases showed that annually 600 million cases are attributed to 31 etiological agents from 2007 to 2015. Approximately 51 million people are infected with Shigella (15). Shigella flexneri is the most common species isolated worldwide (16) and accounts for 65.9% of all cases of shigellosis in a multicenter global study (17). One of the main challenges related to this pathogen is its very low infectious dose; in humans, approximately 10-100 Shigella cells can cause shigellosis. However, 9 × 108 organisms are required to produce shigellosis in rats, which indicates that humans are highly sensitive to this pathogen (18).
Shigella flexneri has a relatively stable genome and acquired the virulence plasmid (Virulence plasmid pINV) early in its evolution (19). The majority of the important virulence factors involved in the Shigella life cycle are localized to a 30 kb region termed the “entry region” on the pINV plasmid (20). This region encodes the type 3 secretion system (T3SS) and Invasion plasmid antigen (Ipa) proteins, which play a key role in initiating the invasion process and pathogenesis of Shigella (21, 22). Ipa proteins are identified as key virulence factors for Shigella infection. The T3SS injects effector proteins into host cells to manipulate the host cell machinery for the benefit of the bacterium (22). At the tip of the T3SS apparatus (T3SA) needle, the IpaD effector protein is polymerized as a homopentamer and is necessary for the control of virulence and secretion of effector proteins (23). Studies have shown that deletion of ipaD not only leads to excessive secretion of other effector proteins but also results in a non-invasive Shigella phenotype (24).
The ipaD gene is highly conserved among Shigella serotypes (ipaD: 96%) (25). The pINV plasmid contains other important genes located outside the “entry region”. The ipaH gene family plays a key role in the pathogenesis of Shigella and is responsible for post-invasion events (15). For example, the IpaH protein encoded by ipaH 7.8 gene helps induce pyroptosis and escape of Shigella from the macrophage phagosome (26, 27). Cultivation of food is one of the main ways to detect and control microbial load, but when the bacteria enter the VBNC state, this method loses its effectiveness (28). In recent years, concerns have been raised about the possibility of regeneration and pathogenicity of VBNC bacteria. Many foods are stored for extended periods in the refrigerator (4°C), and contamination of these foods with the VBNC form of pathogens can pose risks to consumers.
2. Objectives
The purpose of this study was to investigate the possibility of S. flexneri entering the VBNC state under osmotic and nutritional stresses at 4°C.
3. Methods
3.1. Bacterial Strain
Shigella flexneri with PTCC number 1865 (ATCC 12022) was purchased from the Iranian Research Organization for Science and Technology (IROST).
3.2. Viable but Non-culturable Induction
To investigate the possibility of S. flexneri entering the VBNC state, different salt (NaCl) solutions with concentrations of 0, 5, 10, 15, 20, 25, and 30% were prepared. A suspension of S. flexneri with a turbidity of 0.5 McFarland was prepared, and 100 μL of the suspension was added to 4.9 mL of each salt solution, then incubated at 4°C. Each experiment was performed in six replicates. Cultivability was tested by daily inoculation (one loop) of each solution onto BHI (Merck; Germany) medium enriched with 0.6% yeast extract and 1% sodium pyruvate. Inoculated media were incubated at 37°C for 16 - 24 hours. Cultivation of the solutions continued until no bacterial colonies were observed on the BHI medium, which was considered the point at which the bacteria had potentially entered the VBNC state.
3.3. Reverse Transcriptase (RT)-PCR Method
Considering that the half-life of bacterial mRNA is 3 to 5 minutes, the expression of mRNA in the non-cultivable state indicates the survival of bacteria and their entry into the VBNC state (29). Therefore, we investigated the presence of ipaH and ipaD gene mRNA. To achieve this, 1.5 mL of each solution containing VBNC bacteria was centrifuged at 7000 rpm for 5 minutes. The supernatant was removed, and the bacterial pellets were washed three times at 7000 rpm for 5 minutes.
3.3.1. RNA Extraction and cDNA Synthesis
RNA was extracted from the pellets using the DENA Zist Asia RNA extraction kit (Iran) according to the manufacturer's instructions. cDNA was synthesized using the DENA Zist Asia cDNA synthesis kit (Iran), following the manufacturer's recommendations.
3.3.2. PCR Assay
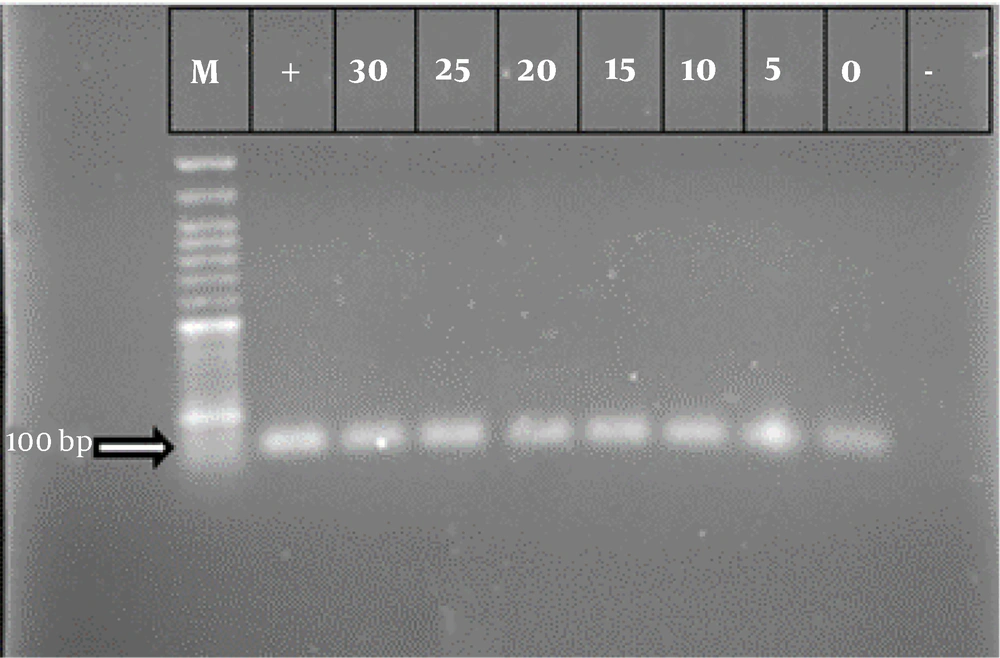
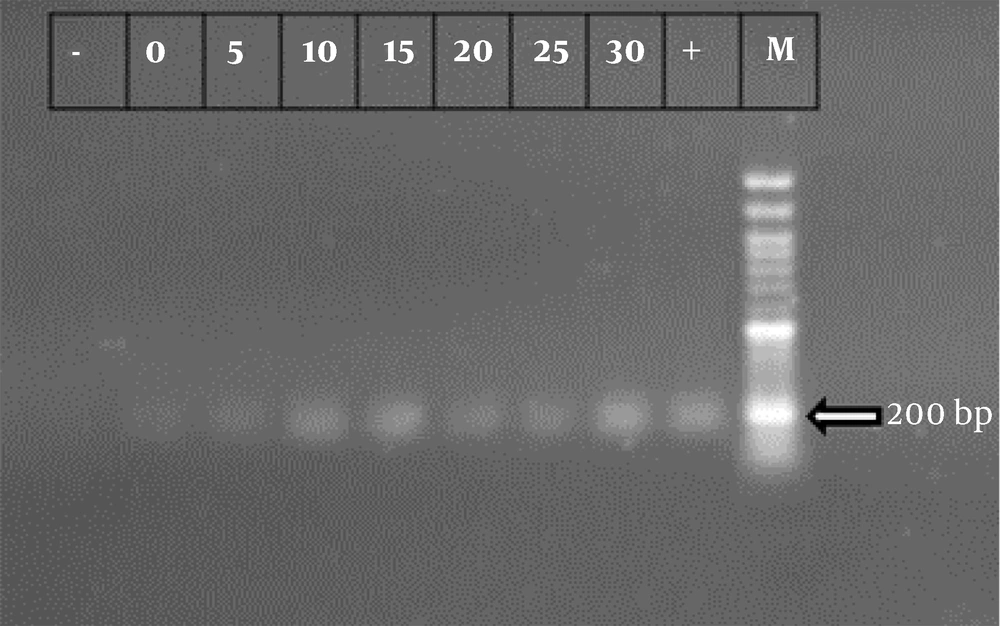
A PCR assay was performed to detect ipaD and ipaH. The primer sequences used in this assay are provided in Table 1. PCR amplification was carried out in a 20 μL reaction mixture containing 5 - 7 ng of template cDNA, 10 μL Master Mix (Ampliqon; Denmark), 0.3 μM of each forward and reverse primer, and distilled water up to 20 μL. The reaction conditions for both genes were as follows: Initial denaturation at 95°C for 4 minutes, followed by 34 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds, extension at 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. After amplification, the products were analyzed by electrophoresis on a 2% agarose gel.
| Primer Name | Sequence 5'→ 3' | Tm | Product Length (bp) |
|---|---|---|---|
| ipaH | 58 | 95 | |
| F | GTTGCTGCTGATGCCACTG | ||
| R | CCTTCTGATGCCTGATGGAC | ||
| ipaD | 58 | 206 | |
| F | CGCTGTTCTTTCCAGTCTTGC | ||
| R | CCTTGCCGATTGTTCCACCT |
3.4. Fluorescent Staining Assay
To evaluate the entry of S. flexneri into the VBNC state using a fluorescence microscope, the non-cultivable bacterial samples were centrifuged at 13,000 rpm for 10 minutes and washed three times with PBS. Untreated bacteria served as the positive control, while bacteria treated with 70% ethyl alcohol were used as the negative control. After resuspending the bacterial pellets in 100 μL of PBS, 10 μL of bacteria were stained with 5 µL of fluorescent dyes, including 0.05 mg/mL acridine orange (AO, Merck) and 0.05 mg/mL propidium iodide (PI, Merck), in PBS under dark conditions for 20 minutes. Smears were prepared from the stained bacteria and observed under a fluorescence microscope (Olympus, Japan).
3.5. Flow Cytometry Assay
To evaluate the entry of S. flexneri into the VBNC state using a flow cytometry assay, the non-cultivable samples were centrifuged at 13,000 rpm for 10 minutes and washed three times with PBS. S. flexneri in the logarithmic phase were used to prepare negative and positive control samples, with the negative control containing live bacteria and the positive control containing dead bacteria (in terms of PI dye absorption and red fluorescence emission). Untreated bacteria served as the negative control, while bacteria treated with 70% ethyl alcohol were used as the positive control. The bacterial pellets were then resuspended in 100 µL of PBS, and 0.5 µL of PI were added to the cells and incubated at room temperature for 20 minutes in the dark. Samples were analyzed using a flow cytometer (BD FACSCalibur; USA) with a single laser emitting excitation light at 488 nm (30).
4. Results
4.1. Non-cultivable State
After the cultivation of S. flexneri in different concentrations of salt solutions, the time at which the bacteria became non-cultivable was observed at salt concentrations of 0, 5, 10, 15, 20, 25, and 30%, respectively, on days 19, 20, 21, 22, 22, 24, and 24. As the salt concentration increased, the time to non-cultivability was delayed.
4.2. RT-PCR
RT-PCR was performed to differentiate the non-cultivable state from bacterial death. For all treatments, the presence of mRNA related to the ipaH and ipaD genes was observed, indicating that the bacteria had entered the VBNC state (Figures 1 and 2).
4.3. Fluorescence Microscopic Results
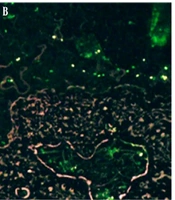
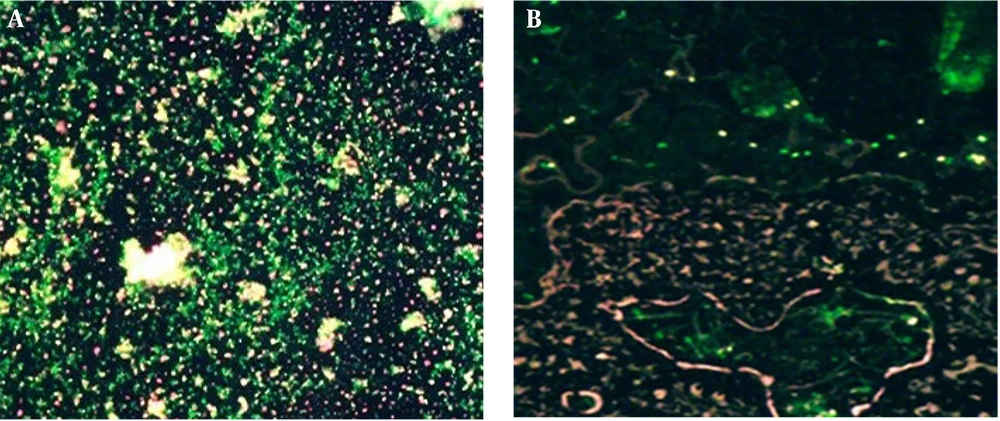
Fluorescent staining of the samples on the 33rd day indicated the presence of living cells. In this case, living cells take up acridine orange (AO) and appear green, while dead cells take up propidium iodide (PI) and appear red. The presence of green cells confirms that S. flexneri has entered the VBNC state (Figure 3).
4.4. Flow Cytometry Results
After S. flexneri became non-cultivable in the investigated treatments, flow cytometry was used to evaluate the bacteria's entry into the VBNC state. The analysis of the flow cytometry data revealed the presence of living S. flexneri cells on the 26th and 96th days of exposure to different salt concentrations (Table 2). These findings also indicated that the number of viable S. flexneri cells was higher in the presence of salt compared to when salt was absent.
| Sample and Marker | 26th Day | 96th Day | ||
|---|---|---|---|---|
| Events | % Gated | Events | % Gated | |
| Positive control | ||||
| All | 10000 | 100.00 | 10000 | 100.00 |
| M1 | 3882 | 38.82 | 4106 | 41.06 |
| NaCl 0% | ||||
| All | 3472 | 100.00 | 475 | 100.00 |
| M1 | 970 | 27.94 | 1 | 0.21 |
| NaCl 5% | ||||
| All | 1031 | 100.00 | 198 | 100.00 |
| M1 | 66 | 6.40 | 0 | 0.00 |
| NaCl 10% | ||||
| All | 1967 | 100.00 | 169 | 100.00 |
| M1 | 132 | 6.71 | 0 | 0.00 |
| NaCl 15% | ||||
| All | 2870 | 100.00 | 26 | 100.00 |
| M1 | 222 | 7.74 | 0 | 0.00 |
| NaCl 20% | ||||
| All | 3837 | 100.00 | 15 | 100.00 |
| M1 | 303 | 7.90 | 0 | 0.00 |
| NaCl 25% | ||||
| All | 1564 | 100.00 | 8 | 100.00 |
| M1 | 148 | 9.46 | 0 | 0.00 |
| NaCl 30% | ||||
| All | 3625 | 100.00 | 53 | 100.00 |
| M1 | 268 | 7.39 | 1 | 1.89 |
| Negative control | ||||
| All | 6235 | 100.00 | 10000 | 100.00 |
| M1 | 112 | 1.80 | 176 | 1.76 |
5. Discussion
After decades of research, the VBNC state has been recognized as an important bacterial survival strategy. A comprehensive understanding of the VBNC state and its underlying mechanisms is critical for ensuring the safety of drinking water and food. This study investigated the survival and viability of S. flexneri PTCC 1865 (ATCC 12022) under osmotic stress in nutrient-depleted conditions at 4°C, as food and water are typically stored at this temperature and may become contaminated from other contents in the refrigerator. The findings of this study indicated that S. flexneri is capable of surviving for extended periods in high NaCl concentration solutions at 4°C. Eventually, it enters the VBNC state and remains in this state for a prolonged duration.
The expression of the ipaD and ipaH genes, which are crucial for the pathogenicity of S. flexneri, following the bacteria's loss of cultivability, suggests their transition into the VBNC state. Moreover, the expression of ipaD in the VBNC state indicates the presence of the virulence plasmid pINV and the retention of pathogenic potential. Additionally, flow cytometry and microscopy results indicate that S. flexneri gradually transitions into the death phase of growth, even when exposed to high salt stress. Shigella flexneri can remain in the VBNC state for an extended period before entering the death phase. The results of the present study are consistent with those of other studies on this subject.
Oliveira et al. reported that S. flexneri 2a does not completely lose cultivability under long-term induction; however, a portion of its population enters the VBNC state. They found that short-term induction resulted in the rapid entry of S. flexneri cells into the VBNC state. They also suggested that low temperatures (4°C) used in nutritional and osmotic stresses can induce the VBNC state while allowing cells to maintain cultivability and viability for a longer period (31). In another study, researchers detected significant quantities of VBNC pathogenic bacteria, including Shigella sp., Escherichia coli, Salmonella sp., Enterococcus faecalis, and Pseudomonas aeruginosa in full-scale drinking water treatment plants. They emphasized that traditional culture-based methods are insufficient for detecting VBNC bacteria (32). Islam et al. (1993) observed that after inoculation of S. dysenteriae type 1 into laboratory microsomes, the bacteria became uncultivable after 2 - 3 weeks, while they entered the VBNC state and remained alive until the sixth week after the start of the study (33).
Islam et al. demonstrated that S. flexneri survived at pH 7.0 and 4°C in 0, 0.5, and 1% salt solutions for 27, 39, and 34 days, respectively (34). In another study, researchers revealed that after inoculating S. dysenteriae type 1 on some inanimate objects, the bacteria entered the VBNC state after 1.5 - 4 hours and remained detectable in this state for five days (35). Rahman et al. (1996) showed that S. dysenteriae type 1 can express the stx gene after entering the VBNC state, and the synthesized Shiga toxin retains its biological activity (36). Scientists explained that the activation of the type II toxin-antitoxin (TA) system in Shigella could partially maintain cell viability in the VBNC state. The TA system halts cell growth and helps Shigella persist under various mild stresses, such as osmotic, temperature, and nutritional stresses (37).
5.1. Conclusions
The results of the present study revealed that S. flexneri could survive and enter the VBNC state under nutritional and osmotic stresses at low temperature (4°C). In this state, although the bacteria are alive, they cannot be detected by conventional culture-dependent methods. The infectious dose of the enteropathogen S. flexneri is very low. The entry of S. flexneri into the VBNC state, coupled with the expression of the ipaD and ipaH genes and the presence of the virulence plasmid pINV, suggests that the bacteria retain their pathogenicity in this state. Furthermore, if the bacteria resuscitate from the VBNC state, it poses a potential risk to food safety and public health.