1. Background
Staphylococcus aureus species are frequently encountered in the medical community, and are considered as an important cause of hospital-acquired infections. Today, the most important issue with S. aureus is that the isolates are getting increasingly methicillin-resistant (1, 2). Treatment options for the infections caused by methicillin-resistant Staphylococcus aureus (MRSA) are limited due to multiple drug resistance (3). Vancomycin, linezolid, and daptomycin are antimicrobial agents used to treat MRSA infections, but the decreased susceptibility of these bacteria to vancomycin in recent years created challenges. Methicillin-resistant S. aureus infections in hospitals lead to higher costs, because they mandate increased antibiotic use and longer hospitalizations; however, the threat that MRSA poses to public health in terms of increased mortality and epidemic potential is of greater importance (4, 5). Due to these reasons, rapid diagnostic tests are important to prevent and treat infectious diseases. Rapid differentiation between MRSA and methicillin-susceptible Staphylococcus aureus (MSSA) is necessary to optimize treatment and minimize costs (6). Making the correct choice of antibiotics based on the antimicrobial susceptibility of MRSA isolates which can be a problem, especially in nosocomial and community-acquired infections may help to reduce the resistance problem by reducing the over-prescription of ineffective antibiotics (7).
The detection of methicillin resistance is always a problem in routine bacteriology laboratory. Various culture methods and assays are performed to identify methicillin resistance including the disk diffusion, E-test, broth microdilution, chromogenic agar medium, oxacillin agar medium, the detection of the mecA gene by polymerase chain reaction (PCR), and latex agglutination assay based on the detection of mecA product (PBP2a) (8, 9). The gold standard to detect the mecA gene is PCR, but most hospitals (apart from advanced central health institutions) do not have laboratory facilities equipped with molecular or phenotypic methods necessary for rapid and accurate identification of methicillin-resistant isolates (10). Thus, there is a clear need to develop simple, quick, and effective methods.
Today, based on the results of former studies, the cefoxitin disk diffusion test is widely used in clinical microbiology laboratories. According to the clinical and laboratory standards institute (CLSI), the cefoxitin disk assay could be performed to demonstrate mecA-mediated oxacillin resistance. Other commonly applied methods are cefoxitin screening with an automated identification system such as VITEK®-2 (bioMerieux SA, Marcy l’Etoile, France) or the BD Phoenix™ (BD Diagnostics, Franklin Lakes, NJ, USA), and the determination of oxacillin minimum inhibitory concentrations (MICs) (11-14). However, after starting the process of sampling, it takes at least 2 days to carry out bacterial cultivation, identification, and antibiotic-susceptibility testing. Therefore, tests that can make MRSA identification faster are required in order to initiate effective antibiotic treatments earlier.
2. Objectives
The newly developed StaResMet® kit is based on a microdilution method, and allows the determination of cefoxitin MIC in 6 hours. Cefoxitin MICs in the current study were determined using a colorimetric read out, where the color change occurred in the wells of the kit in which there was bacterial growth. The current study aimed at assessing the effectiveness of the StaResMet® kit in the rapid detection of methicillin resistance S. aureus species isolated from clinical samples.
3. Methods
3.1. Bacterial isolates
3.1.1. Phase I
In this phase, the test was performed on frozen stocks. A total of 189 MRSA and 177 MSSA species isolated from various clinical samples were analyzed using the StaResMet® test kit at 3 centers: the department of medical microbiology in the school of medicine at Adnan Menderes University; the department of medical microbiology in the school of medicine at Canakkale Onsekiz Mart University; the training and research hospital microbiology laboratory at Ahi Evran University. Out of 189 MRSA samples evaluated in the current study, 100 species were mecA-positive based on polymerase chain reaction (PCR) assay at Adnan Menderes University. The MICs of oxacillin for 47 of the isolates were determined to be at least 4 µg oxacillin mL-1 using the BD Phoenix™ (BD Diagnostics, Franklin Lakes, NJ, USA) at Canakkale Onsekiz Mart University. Cefoxitin-screening using a VITEK®-2 (bioMérieux SA, Marcy l’Etoile, France) at Ahi Evran University validated 42 of the isolates as MRSA. Using automated systems, Canakkale Onsekiz Mart University identified 49 of the MSSA isolates, and Ahi Evran University identified 128 of the MSSA isolates. In the study, S. aureus ATCC 29213 (methicillin-susceptible) and ATCC 43300 (methicillin-resistant) were used as controls.
3.1.2. Phase II
In this phase, S. aureus species isolated in daily routine practices were tested. Application of the test at the same time and obtaining the results on the same day were evaluated. A total of 28 MRSA and 75 MSSA species isolated from clinical specimens at Microbiology laboratory of school of medicine at Samsun Ondokuz Mayis University were included. In this phase, suspected colonies of S. aureus were tested to evaluate culture plates. Preliminary identification was made according to colony morphology, Gram staining, and catalase reaction. And isolates were identified by Vitek MS (bioMerieux SA, Marcy l’Etoile, France) automated system. After the identification of the isolates within minutes, isolates were tested with StaResMet® kit. Antimicrobial susceptibilities of the isolates were determined by Vitek2 Compact (bioMérieux SA, Marcy l’Etoile, France) system.
3.2. StaResMet® Kit Assays
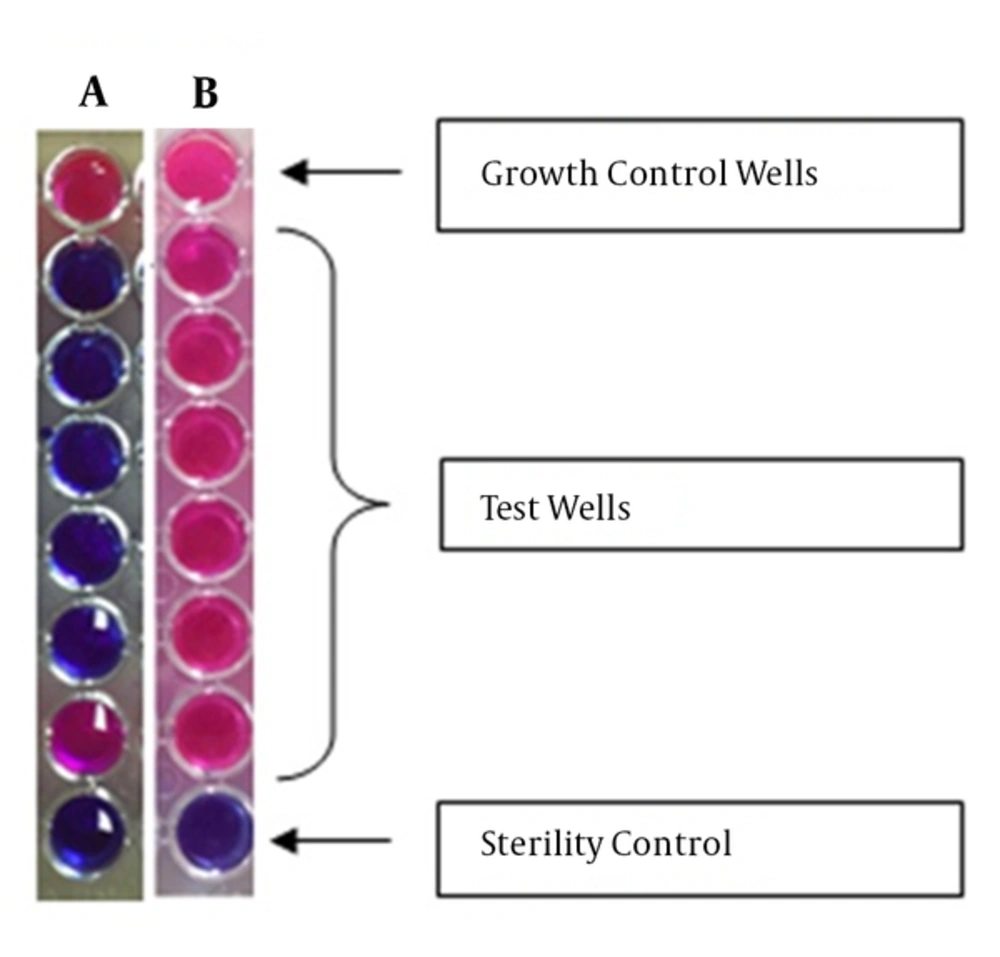
The StaResMet® kit assay was performed according to the manufacturer’s protocol. The kit includes 2 solutions (1 and 2), and a U-bottom 96-well plate with wells containing various amounts of cefoxitin. The wells in row A did not contain antibiotics and were used as growth controls, and the wells in row H did not contain both bacterial suspensions and antibiotics and were used as sterility controls. After addition of bacteria and growth media, rows B, C, D, E, F, and G contained 32, 16, 8, 4, 2, and 1 µg/mL cefoxitin, respectively. Each plate allowed for testing 12 bacterial isolates, with 8 wells (A1-H1, A2-H2, etc.) used for each isolate (Figure 1). First, Solution 1 was added to all wells. Bacterial suspensions with 0.5 and 1 McFarland standards were prepared from freshly grown (i.e., grown overnight) bacterial cultures. Every well, except for those in row H, was inoculated with 10 µL of bacterial suspension. Plates were, then, incubated at 35˚C for 5 hours. After the incubation, 30 µL of Solution 2 (the blue indicator solution) was added to each well. Plates were, then, incubated at 35°C for 1 hour. A successful test indicated the appearance of a red color in the growth control well. Among antibiotic-containing wells, the last well in which a blue color was observed indicated the MIC. To determine methicillin resistance, an MIC of at least 8 µg/mL cefoxitin was taken as a threshold, in accordance with the European committee on antimicrobial susceptibility testing (EUCAST) Clinical Breakpoint version version 5.0, and the CLSI Performance Standards for Antimicrobial Susceptibility Testing; 24th informational supplement (M100-S24).
Images of the wells from 2 investigated assays are shown. A, the results indicated the MIC of 2 µg/mL for cefoxitin; B, The results indicated the MIC of > 32 µg/mL for cefoxitin. The top wells in A and B are the growth control wells with no cefoxitin and the bottom well contained no bacteria. Red color indicates bacterial growth, while blue color indicates no bacterial growth. Concentrations of cefoxitin (from top to bottom, excluding control wells) were 32, 16, 8, 4, 2, and 1 µg/mL, respectively.
4. Results
4.1. Phase I
The distribution of cefoxitin MICs obtained in the first phase of the current study using the StaResMet® kit with S. aureus isolates is shown in Table 1. In the current study, the identity of all 189 previously characterized MRSA isolates were confirmed using the StaResMet® kit. For the MRSA isolates, cefoxitin MICs were determined > 32 µg/mL for 132 isolates, 32 µg/mL for 46, 16 µg/mL for 4, and 8 µg/mL for 7 isolates. Likewise, all 177 MSSA isolates were identified as MSSA using the StaResMet® kit, and cefoxitin MICs for these isolates ranged from 1 to 4 µg/mL (Table 1). In this phase, specificity and sensitivity were 100%.
Cefoxitin MICs Determined by StaResMet® Kit
4.2. Phase II
The distribution of cefoxitin MICs is shown in Table 1. All of the results were obtained in the same shift by StaResMet® kit. In this phase of the study, cefoxitin MIC value of 2 of the 28 MRSA isolates determined by Vitek-2 Compakt system were 4 µg/mL in StaResMet® kit. These isolates were also tested with broth microdilution method and the MIC values were 4 µg/mL similar to StaResMet® kit. For the MRSA isolates, cefoxitin MICs were > 32 µg/mL for 1 isolate, 32 µg/mL for 3 isolates, 16 µg/mL for 18 isolates, and 8 µg/mL for 4 isolates (Table 1). In this phase, specificity and sensitivity were 100%. Therefore, results of the current study experiments obtained by the StaResMet® kit were in perfect concordance with those obtained using well established clinical diagnostic methods.
5. Discussion
Methicillin-resistant S. aureus is one of the most common causes of community-acquired and nosocomial infections, both in Turkey and worldwide. When MRSA is detected in hospitalized patients, especially the ones treated in the intensive care units (ICUs), appropriate treatment and infection-control precautions should be started immediately (4, 15). Though identifying S. aureus is not difficult, phenotyping methods in order to detect methicillin resistance is time-consuming. Methicillin resistance occurs as a result of the synthesis of PBP2a, a penicillin-binding protein encoded by the mecA gene. Although detecting the mecA gene by molecular methods is the gold standard, these methods cannot be performed in most laboratories (10, 16). For this reason, there is still a need to develop new assays as alternatives to the phenotyping methods currently in use.
Rapid detection of resistance genes or phenotypes in microorganisms was investigated in several studies in recent years (17-19). The commonly used commercial kits based on molecular methods include the BD GeneOhmTM MRSA ACP assay, the GeneXpert® MRSA assay, the Hyplex Staphylo Resist test system, and the BD GeneOhmTM StaphSR assay (5, 7, 20). However, these molecular methods cannot be used in most clinical microbiology laboratories in Turkey due to their high costs and lack of required technical equipment. Other options include the PBP2a latex agglutination test and identification with chromogenic agar medium. While MRSA identification is performed with automated systems such as the VITEK®-2 or the BD Phoenix™ in most clinical microbiology laboratories, cefoxitin disk diffusion test and commercial kits that detect coagulase production are used to identify MRSA in some smaller hospitals.
Obtaining an antibiogram by these methods requires at least 2 days from the time of sampling, during which the initial culture, subsequent passage, and characterizations are carried out. Therefore, rapid and accurate detection is extremely important to save time in hospitals with less robust facilities, especially for life-threatening infections involving sepsis (21). Considering these factors, the StaResMet® test is very promising. The test determines the MIC of cefoxitin against a bacterial isolate based on culturing the isolate with a range of drug concentrations and providing a colorimetric indication of bacterial growth. The application of this method in clinical microbiology laboratories is very practical and easy, and allows the evaluation of methicillin resistance only 6 hours after the identification of S. aureus (after Gram staining and coagulase testing). Several studies compared methods that rely on liquid microdilution and colorimetric to determine oxacillin and vancomycin resistance in S. aureus isolates, and colorimetric methods are also used to detect antibiotic resistance in Mycobacterium tuberculosis (22-25).
In one study, when a colorimetric nitrate reductase test and the Resazurin microplate method for the rapid detection of MRSA were compared to reference methods, categorical (agreement of interpretive results between a new device under evaluation and a standard reference method) and essential (agreement within plus or minus, one two-fold dilution of the new device under evaluation with the reference method MIC determination) agreements among the methods were 100% and 99.6%, respectively (26). Therefore, the existing research showed that these easy-to-apply methods give fast and reliable results. The findings of the current study using the StaResMet® kit were in full agreement with these previously published reports.
Soysal et al. (27) evaluated StaResMet® for the rapid detection of methicillin resistance in S. aureus species. In their study, 118 MRSA and 159 MSSA isolates were tested by the broth microdilution method (the reference method), Vitek-2 Compakt system and StaResMet®. They found that all results were concordant and their specificity and sensitivity were 100%. According to the results of the current study, StaResMet® kit provides daily methicillin susceptibility results after the identification of the isolate with no wait for a day of incubation. Furthermore, the method can prevent unnecessary use of vancomycin and teicoplanin and provides early and appropriate treatment of patients with severe infections.
6. Conclusions
Overall, it was concluded that the StaResMet® test kit was very accurate, rapid, and practical for routine use in clinical microbiology laboratories. Importantly, this diagnostic kit had the potential to facilitate faster and more effective application of antibiotics to treat MRSA.