1. Introduction
Aspergillus genus is one of the most common causes of pulmonary fungal infections with high morbidity and mortality worldwide, followed by Cryptococcus neoformans and Pneumocystis jirovecii (1). The incidence of aspergillosis has increased significantly (2, 3), mainly in immunodeficient patients with hematologic malignancies, solid organ transplantation, and corticosteroid therapy (4). Coronavirus disease 2019 (COVID-19) has been declared a pandemic since the end of December 2019, leading to millions of respiratory emergencies. The prevalence of COVID-19 may be highly associated with an increased risk of pulmonary aspergillosis, possibly due to disrupted immune systems (5, 6). As reported, it was not considered as common in COVID-19 patients, which may be reflected by the incidence rate of COVID-19-associated pulmonary aspergillosis (CAPA) (7) of less than 1%. Aspergillus latus, a newly rare species that arose from allodiploid hybridization between A. spinulosporus and a proximate species of A. quadrilineatus, shows heterogeneity in infection-related traits compared to its parent species. For example, A. latus spores are more resistant to antifungals and oxidative stressors (1). Understanding of A. latus remains limited (2). It has never been reported in cases of pulmonary aspergillosis, let alone co-infections in COVID-19 patients (8).
Owing to nonspecific clinical manifestations and radiographic evidence, it is difficult to distinguish aspergillosis from COVID-19 alone, resulting in delayed diagnosis and lack of appropriate treatment (3, 4). Therefore, timely diagnosis and antifungal treatment of COVID-19-associated pulmonary aspergillosis is essential for patient relief. Growing evidence has shown the tremendous potential of metagenomic next-generation sequencing (mNGS) in clinical applications due to unbiased identification and rapid detection (5). Based on bronchoalveolar lavage fluid (BALF) samples, mNGS was employed to assist clinical infection-related diagnosis. Briefly, the mNGS process includes sample pretreatment, nucleic acid isolation (6), library construction and sequencing (7), bioinformatic analysis, and interpretation (8, 9). For each run, parallel human nucleic acid as a negative control was prepared and detected using the same protocol to calculate background and monitor sample-to-sample contamination (6). Here, based on mNGS, we identified co-infections of A. latus and P. jirovecii as the causes of fatal pneumonia in an immunocompromised COVID-19 patient.
2. Case Presentation
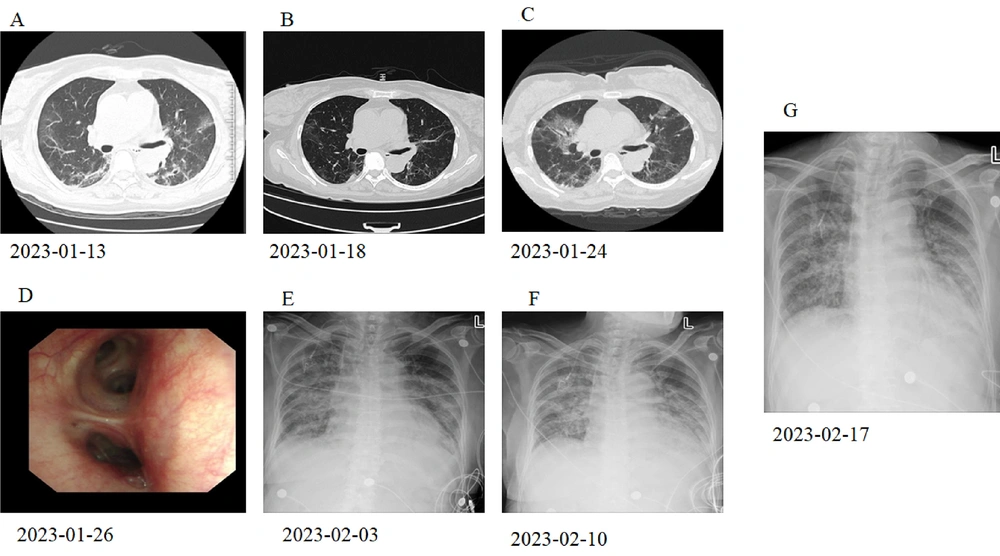
A 67-year-old female with a history of diffuse large B cell lymphoma was receiving a combination of obinutuzumab and gemcitabine plus oxaliplatin (GemOx). The patient was admitted to our hospital complaining of a 10-day discomfort of fever and cough. She was initially diagnosed with severe COVID-19 pneumonia and non-Hodgkin's lymphoma (NHL). After one week of treatment with oxygen inhalation, nematavir/ritonavir tablets (Paxlovid™), methylprednisolone, moxifloxacin, and anticoagulation, her body temperature remained normal, and partial absorption of lesions in the lung was also observed through chest CT scans (Figure 1A and B). Methylprednisolone (12 mg/day) was continued after her discharge. However, the patient was readmitted with a temperature of 37.9°C, complaining of a four-day history of fever, paroxysmal cough with little phlegm, and chest tightness. She had clear consciousness, a normal heart rhythm, and moist rales in her chest. The chest CT showed multiple inflammatory features throughout both lungs, indicating exacerbation of lesions in the lungs (Figure 1C).
Radiographic images of the patient alongside therapy. A, the chest CT at external hospital showed scattered exudative lesions in both lungs; B, the chest CT showed that the partial absorption of lesions in both lungs; C, the chest CT showed multiple inflammation in both lungs, which was advanced; D, fiberoptic examination exhibited no obvious abnormality; E, digital radiography examination showed the infection of extensive exudation in both lungs; F, digital radiography examination showed the infection of extensive exudation in both lungs; G, digital radiography examination showed the infection of extensive exudation in both lungs.
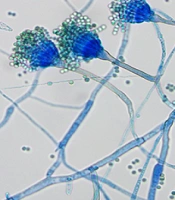
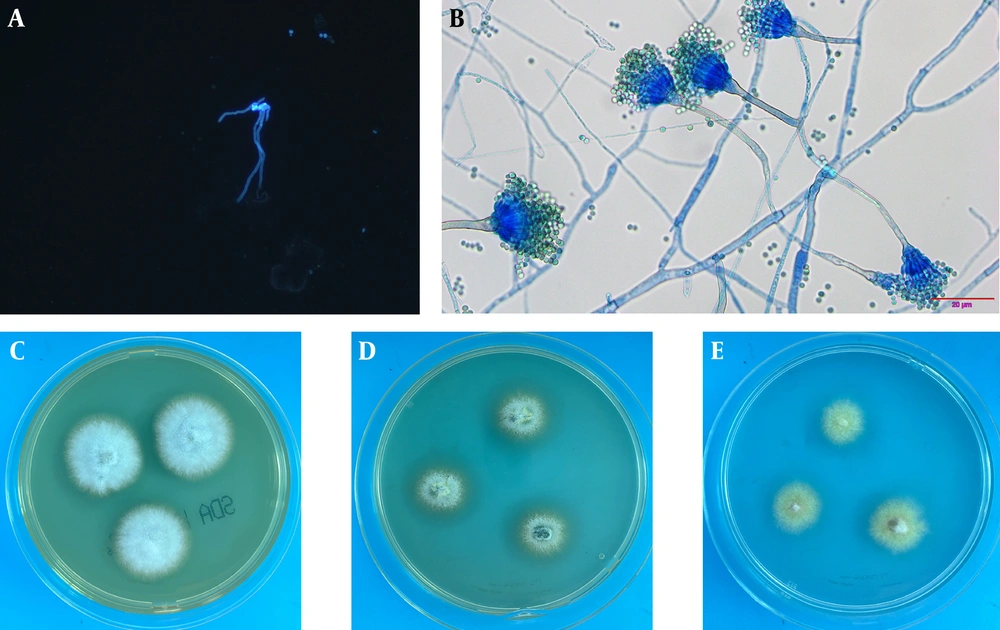
For a definitive diagnosis, fibrobronchoscopy showed an unobstructed lumen with slight mucosal congestion (Figure 1D). Then BALF was collected from the right upper lobe for pathology, microbiological culture, and mNGS. The cytopathology of BALF yielded negative results. Aspergillus-like infection was indicated by the positive result of the galactomannan (GM) test (3.627), and fungal fluorescence staining showed coarse, divided, and bifurcated hyphae (Figure 2A). mNGS further confirmed co-incidence of A. latus (415 reads), P. jirovecii (4 reads), and human herpes virus type 1 (1 read) in the patient with SARS-CoV-2 infection (Table 1). After that, the villous isolate of A. latus from BALF was obtained at 27°C on selective medium, including Sabouraud dextrose agar (SDA), Czapek Dox agar (CZA), and potato dextrose agar (PDA) plates (Figure 2B-D). Under lactophenol cotton blue (LPCB) staining, yellow-green conidia were radially arranged at the top of the vesicle (Figure 2E).
The patient with co-infections was treated with a combination of voriconazole (0.2 g, q12h) and Sulfadiazine (2 tablets, q6h). Although a continuous decrease in c-reactive protein was observed, the patient continued to show signs of extensive infection in both lungs (Figure 1E). After one week of treatment with plasma from recovered COVID-19 patients, the test for SARS-CoV-2 turned negative, but poor lung imaging persisted under digital radiography examination (Figure 1F). Subcutaneous injection of thymosin-alpha 1 (Zadaxin™, 1.6 mg/day) and oral nintedanib (150 mg/ 2 days) were used to treat pulmonary infection. Unfortunately, the patient suffered from extensive oral ulcers, increased dyspnea, and agranulocytosis. Chest radiographs at the bedside showed extensive exudation in both lungs (Figure 1G). Eventually, the patient died of respiratory failure 29 days after admission.
Identification of Aspergillus latus from bronchoalveolar lavage fluid (BALF). A, Fungal fluorescence staining showed coarse, divided and bifurcated hypha; B, BALF were cultured at 27°C for 3 days on Sabouraud dextrose agar (SDA) plate to form medium sized, villous opalescent colonies; C, BALF was cultured at 27°C for 3 days on a Czapek Dox agar (CZA) plate, and medium sized, villous faint yellow colonies were formed; D, BALF was cultured at 27°C for 3 days on potato dextrose agar (PDA) plate medium to form medium sized, marginal villous pale green colonies; E, lactophenol cotton blue (LPCB) showed that the conidial head was short columnar, and the conidial peduncle was smooth and curved. Microconidia are spherical and green.
| Genus Name | Relative Abundance, (%) | Sequences | Species Name | Confidence Level, (%) | Sequences |
|---|---|---|---|---|---|
| Aspergillus | 15.5 | 853 | Aspergillus latus | 99 | 415 |
| Pneumocystis | 0.5 | 4 | Pneumocystis jirovecii | 99 | 4 |
a Pathogens are classified into diverse species based on bacteria, fungi, viruses, and parasites, and its relative abundance is the relative proportion of genomes in the corresponding classification of the pathogen. Sequences means high-throughput sequencing sequences that are uniquely aligned to genus or species-specific sequences. Confidence level is defined as the reliability of the pathogen identified in the sample based on sequence alignment.
3. Discussion
Since the outbreak of COVID-19 pneumonia in December 2019, CAPA, a secondary pulmonary Aspergillus infection, has been detected in patients with severe COVID-19, which is a life-threatening condition for the immunocompromised (10). Those with hematologic malignancies, solid organ transplantation, and corticosteroid hormone therapy are more likely to develop CAPA (11-13). For the patient we described, the history of radiotherapy and chemotherapy due to the diagnosis of diffuse large B cell lymphoma, the subsequent destructive lung structure, and the treatment of severe COVID-19 pneumonia with steroids and broad-spectrum antibiotics suggest that she was at high risk for developing CAPA due to her immunocompromised status.
Aspergillus is widely distributed in environmental samples, such as soil, sanitation, and medical facilities. As reported, A. latus is a newly rare species that evolved from A. spinulosporus and a proximate species of A. quadrilineatus through gene fusion (14). Hybridization may lead to new species acquiring new metabolic pathways, producing new metabolites or virulence factors, and altering invasive ability. It is suggested that the characteristics of A. latus, with unique genetic characteristics, may be different from any of its parent strains (15). These may pose significant challenges to the study of pathogenicity, disease diagnosis, and treatment.
It is challenging to distinguish pulmonary aspergillosis from COVID-19 pneumonia alone due to the similarity in clinical manifestations and radiographic evidence (3, 4). The most common method of diagnosing Aspergillus is to isolate the fungi from BALF or tracheal aspirate in culture, though the accuracy of microbiological diagnosis depends largely on laboratory experience and conditions. Timely and precise identification of pathogens based solely on traditional culture is not practical (16). Other diagnostic tests, such as GM, could only identify Aspergillus infections at the genus level (17). Compared to those techniques, mNGS has proved to be a promising clinical diagnostic tool, providing unbiased, culture-free, time-saving means for detecting all potential pathogens, especially for intractable diseases (5, 18). In this case, the pathogens from BALF were not recovered on culture; subsequent GM tests revealed Aspergillus infection, and mNGS further demonstrated co-infections of A. latus and P. jirovecii in COVID-19 patients, indicating the promising complementary role of mNGS in diagnosing Aspergillus infections.
Unfortunately, the patient died due to delayed antifungal treatment for CAPA. On admission, the patient received methylprednisolone for the treatment of SARS-CoV-2 infection. Corticosteroids and cytokine blockers (Tocilizumab™) are reported to inhibit residual immune function and increase viral cell lysis capacity, further accelerating fungal invasion and infection (12, 19, 20). An earlier diagnosis of aspergillosis may have had an important impact on treatment and prognosis (21). Here, a high level of awareness should be exercised for CAPA, and corticosteroids should be used with caution in the clinical setting, especially for severe COVID-19 patients.
5.1. Conclusions
A fatal co-infection of A. latus and P. jirovecii in a COVID-19 positive patient was determined by mNGS for the first time. Clinicians should be aware of the risk of CAPA, especially in patients with severe COVID-19. In situations where CAPA is known to occur, samples such as BALF should be collected for continuous monitoring to ensure timely treatment of aspergillosis. Additionally, attention should be given to the emergence of rare and pathogenic fungi. Quick and accurate detection of the pathogen through mNGS could expedite the diagnostic process and potentially reduce patient mortality.