1. Background
Carbapenem antibiotics are currently the most widely used and potent β-lactam antibiotics, with the broadest antibacterial spectrum, often referred to as the last line of defense against Gram-negative bacteria. However, due to their frequent use in clinical practice, many Enterobacteriaceae have developed resistance to them. Carbapenem-resistant Enterobacteriaceae (CRE) refers to a group of Enterobacteriaceae that are resistant to any carbapenem antibiotic or capable of producing carbapenemases (1). Carbapenem-resistant Enterobacteriaceae is classified at the highest priority level on the WHO's list of antibiotic-resistant bacteria (2). Without effective intervention measures, CRE could become widespread in healthcare institutions within the next ten years, potentially developing into an endemic issue (3). Bloodstream infections caused by CRE are associated with alarmingly high mortality rates, ranging from 30% to 80% (4). The severe drug resistance posed by CRE may result in limited or no effective treatment options. Additionally, significant geographical differences exist in the epidemiological distribution and multilocus sequence typing (MLST) of CRE.
2. Objectives
This study conducted an epidemiological analysis and MLST typing of carbapenem-resistant Klebsiella pneumoniae (CRKP) strains isolated from clinical samples in hospitals from 2020 to 2022. The aim was to understand the transmission and epidemiological characteristics of CRE, ultimately providing evidence for the prevention and control of CRE infections and improving clinical anti-infection treatment strategies.
3. Methods
3.1. Basic Information Collection of Carbapenem-Resistant Enterobacteriaceae Infected Individuals
Based on drug sensitivity results, 187 patients with CRE strains isolated from the First Affiliated Hospital of Guizhou University of Traditional Chinese Medicine between 2020 and 2022 were screened (excluding patients with duplicate CRE strains) and included in the study. Relevant information on the infected individuals was collected. Inclusion criteria for CRE-infected patients excluded those with incomplete medical records and test results. Each patient with the same hospitalization number and CRE bacteria was enrolled only once. This study was approved by the hospital ethics committee.
3.2. Main Instruments and Reagents
The VITEK 2 Compact fully automated microbial identification analyzer (BioMerieux, France), VITEK MS fully automated microbial mass spectrometry detection system (BioMerieux, France), nutrient agar plates (Dijing, Guangzhou), Simmons citrate agar (HB0115, Qingdao Haibo), inositol (Macklin), meropenem (Macklin), and bacterial DNA extraction kit (Genefist) were used. All reagents were used within their validity period.
3.3. Isolation, Cultivation, Identification, and Drug Sensitivity Experiments of Carbapenem-Resistant Enterobacteriaceae Strains
Microbiological samples submitted for clinical testing were inoculated onto nutrient agar plates and incubated at 36°C for 18 - 24 hours. Bacteria were identified using the VITEK MS fully automated microbial mass spectrometry detection system, followed by drug sensitivity testing with the VITEK 2 Compact fully automated microbial identification analyzer. The results were interpreted according to the standards published by the Clinical and Laboratory Standards Institute (CLSI) in the United States.
3.4. Review Identification and Bacterial DNA Extraction of Carbapenem-Resistant Klebsiella pneumoniae Strains
Resuscitate CRKP strains on a Simmons citrate inositol agar plate containing meropenem at a final concentration of 2 μg/mL (5). Select the colonies growing on the plate and extract bacterial DNA according to the instructions provided with the reagent kit.
3.5. Multilocus Sequence Typing
Referring to the MLST website of the Pasteur Institute (https://bigsdb.pasteur.fr/), Beijing Tianyi Huiyuan Biotechnology Co., Ltd. was commissioned to synthesize the amplification products and sequencing primers for seven housekeeping genes: GapA, infB, mdh, pgi, phoE, rpoB, and tonB. The PCR amplification products were subjected to bidirectional sequencing, and the sequencing results were submitted to the MLST database (https://bigsdb.pasteur.fr/klebsiella/primers-used/). The sequences were aligned, the allele profile was obtained, and the sequence type (ST) of the tested strain was determined.
3.6. Data Analysis and Processing
The data were organized using WHONET 5.6 software, with quantitative variables presented as mean ± standard deviation (SD) and categorical variables expressed as frequency (percentage). Statistical analysis was performed using SPSS version 23.0 for Windows (IBM, Armonk, New York).
4. Results
4.1. General Situation
From 2020 to 2022, the hospital isolated 187 strains of CRE bacteria, including 78 strains in 2020, 58 in 2021, and 51 in 2022. The majority of infections occurred in males, comprising 76.47% (143/187) of the cases. Among the infected individuals, patients over 60 years old accounted for 78.07% (146/187), with those over 80 years old making up 63.70% (93/146) of the total infections in this age group.
4.2. Distribution of Carbapenem-Resistant Enterobacteriaceae Departments and Specimen Sources
A total of 19 departments in the hospital detected CRE, with the ICU wards, geriatric departments, and rehabilitation departments showing higher detection rates, accounting for 35.29% (66/187), 11.23% (21/187), and 8.02% (15/187), respectively. In terms of specimen sources, sputum, clean midstream urine, and secretions were the primary sources, accounting for 43.85% (82/187), 32.09% (60/187), and 13.90% (26/187), respectively. Please refer to Table 1 for details.
| Classification | No. (%) |
|---|---|
| Age (y) | |
| ≤ 35 | 4 (2.14) |
| 35 ~ 60 | 37 (19.79) |
| 60 ~ 80 | 53 (28.34) |
| ≥ 80 | 93 (49.73) |
| Gender | |
| Male | 143 (76.47) |
| Female | 44 (23.53) |
| Specimen source | |
| Sputum | 82 (43.85) |
| Clean midstream urine | 60 (32.09) |
| Secretion | 26 (13.90) |
| Blood | 12 (6.42) |
| Serosal cavity effusion | 3 (1.60) |
| Catheter tip | 1 (0.53) |
| Genital secretions | 2 (1.07) |
| Cerebrospinal fluid | 1 (0.53) |
| Department | |
| Intensive care unit | 66 (35.29) |
| Geriatrics | 21 (11.23) |
| Neurosurgery | 10 (5.35) |
| Urology | 10 (5.35) |
| Rehabilitation | 15 (8.02) |
| Anorectal | 11 (5.88) |
| Hepatobiliary Surgery | 10 (5.35) |
| Neurology | 13 (6.95) |
| Cardiology | 6 (3.21) |
| Emergency | 9 (4.81) |
| Orthopedics | 2 (1.07) |
| Dermatology | 2 (1.07) |
| Respiratory medicine | 5 (2.67) |
| Otolaryngology | 1 (0.53) |
| Gastroenterology | 1 (0.53) |
| Nephrology | 2 (1.07) |
| Traditional Miao Medicine and Pharmacy | 1 (0.53) |
| Oncology | 1 (0.53) |
| Obstetrics | 1 (0.53) |
4.3. Composition of Carbapenem-Resistant Enterobacteriaceae Bacteria
From 2020 to 2022, the primary CRE bacteria isolated in the hospital were carbapenem-resistant K. pneumoniae and Escherichia coli, accounting for 80.21% (150/187) and 14.44% (27/187), respectively, with an additional 3.74% (7/187) comprising other E. coli strains. Please refer to Table 2 for details.
| Bacterial Classification | 2020 | 2021 | 2022 | Subtotal |
|---|---|---|---|---|
| Escherichia coli | 11 (14.10) | 7 (12.07) | 9 (17.65) | 27 (14.44) |
| Klebsiella pneumoniae | 64 (82.05) | 48 (82.76) | 38 (74.51) | 150 (80.21) |
| Strange Proteobacteria | 0 (0.00) | 2 (3.45) | 0 (0.00) | 2 (1.07) |
| Enterobacter cloacae | 3 (3.85) | 1 (1.72) | 3 (5.88) | 7 (3.74) |
| K. oxytoca | 0 (0.00) | 0 (0.00) | 1 (1.96) | 1 (0.53) |
| Subtotal | 78 (100.00) | 58 (100.00) | 51 (100.00) | 187 (100.00) |
a Values are expressed as No. (%).
4.4. Drug Resistance Rate Carbapenem-Resistant Enterobacteriaceae Bacterial Resistance Rate
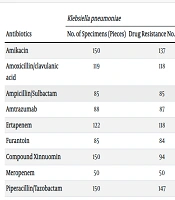
The analysis shows that CRE exhibits high resistance to commonly used antibiotics, including β-lactams, quinolones, nitrofurans, aminoglycosides, and tetracyclines. However, carbapenem-resistant K. pneumoniae has a low resistance rate to tigecycline and cefotaxime/avibactam, at 0% and 4.08%, respectively. Carbapenem-resistant E. coli shows a resistance rate of 0% to tigecycline and 5.41% to amikacin. Additionally, carbapenem-resistant E. coli exhibits 0% resistance to both tigecycline and amikacin. The resistance rates of CRE bacteria to the corresponding antibiotics are presented in Table 3.
| Antibiotics | Klebsiella pneumoniae | Escherichia coli | Enterobacter cloacae | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of Specimens (Pieces) | Drug Resistance No. (Copies) | Drug Resistance Rate (%) | No. of Specimens (Pieces) | Drug Resistance No. (Copies) | Drug Resistance Rate (%) | No. of Specimens (Pieces) | Drug Resistance No. (Copies) | Drug Resistance Rate (%) | |
| Amikacin | 150 | 137 | 91.33 | 27 | 1 | 3.70 | 7 | 0 | 0.00 |
| Amoxicillin/clavulanic acid | 119 | 118 | 99.16 | 23 | 22 | 95.65 | - | - | - |
| Ampicillin/Sulbactam | 85 | 85 | 100.00 | - | - | - | - | - | - |
| Amtrazumab | 88 | 87 | 98.86 | - | - | - | - | - | - |
| Ertapenem | 122 | 118 | 96.72 | 23 | 21 | 91.30 | 3 | 3 | 100.00 |
| Furantoin | 85 | 84 | 98.82 | - | - | - | - | - | - |
| Compound Xinnuomin | 150 | 94 | 62.67 | 27 | 13 | 48.15 | 7 | 4 | 57.14 |
| Meropenem | 50 | 50 | 100.00 | 6 | 6 | 100.00 | - | - | - |
| Piperacillin/Tazobactam | 150 | 147 | 98.00 | 27 | 25 | 92.60 | 7 | 7 | 100.00 |
| Gentamicin | 85 | 79 | 92.94 | - | - | - | - | - | - |
| Tigecycline | 36 | 0 | 0.00 | 24 | 0 | 0.00 | 7 | 0 | 0.00 |
| Cefepime | 150 | 147 | 98.00 | 27 | 27 | 100.00 | 7 | 4 | 57.14 |
| Cefuroxime | 119 | 118 | 99.16 | 23 | 23 | 100.00 | 7 | 7 | 100.00 |
| Cefuroxime axetil | 119 | 118 | 99.16 | 23 | 23 | 100.00 | 7 | 7 | 100.00 |
| Cefoperazone/Sulbactam | 128 | 125 | 97.66 | 27 | 23 | 85.19 | 7 | 7 | 100.00 |
| Ceftriaxone | 150 | 149 | 99.33 | 27 | 27 | 100.00 | - | - | - |
| Ceftazidime | 150 | 146 | 97.33 | 27 | 26 | 96.30 | 5 | 5 | 100.00 |
| Ceftazidime/Avibactam | 98 | 4 | 4.08 | - | - | - | - | - | - |
| Cefotetan | 85 | 84 | 98.82 | - | - | - | - | - | - |
| Cefoxitin | 119 | 118 | 99.16 | 23 | 22 | 95.65 | - | - | - |
| Cefozolin | 95 | 95 | 100.00 | - | - | - | - | - | - |
| Tobramycin | 88 | 82 | 93.18 | - | - | - | - | - | - |
| Imipenem | 150 | 145 | 96.67 | 27 | 24 | 88.89 | 7 | 4 | 57.14 |
| Ciprofloxacin | 88 | 85 | 96.59 | - | - | - | - | - | - |
| Levofloxacin | 150 | 145 | 96.67 | 27 | 24 | 88.89 | 5 | 3 | 60.00 |
a "-" indicates that the antimicrobial drug has not been used for drug sensitivity testing.
4.5. Multilocus Sequence Typing Results
Out of 23 preserved strains of CRKP bacteria in the laboratory, 11 strains survived. CRKP bacterial DNA was extracted, and seven housekeeping genes were amplified using the PCR method. After aligning the sequences, the ST types were identified. The results revealed that two sequence types were detected among the 11 strains, with ST11 being the predominant type, accounting for 90.91% (10/11), and ST15 accounting for 9.09% (1/11).
5. Discussion
Enterobacteriaceae are significant pathogens in both community-acquired and hospital-acquired infections, and carbapenem antibiotics are the most effective treatment for severe Enterobacteriaceae infections. However, the emergence of carbapenem-resistant Enterobacteriaceae poses substantial challenges to clinical treatment, with the growing concern of having no viable therapeutic options. Studies have shown that the incidence and infection rates of CRE in China are steadily rising and surpassing those in other countries. According to data from the 2020 National Antibiotic Resistance Monitoring Network (CHINET), the resistance rate of K. pneumoniae to carbapenems increased from 4.8% in 2014 to 10.5% in 2019 (6). CRE represents the most urgent and threatening drug-resistant bacteria in hospital settings, requiring stringent monitoring and control.
This study revealed that the CRE infection rate at a traditional Chinese medicine hospital primarily affected elderly patients, with those aged ≥ 60 years accounting for 78.07% of the total infection rate. Among them, elderly patients aged ≥ 80 years comprised 63.70%, a finding consistent with research both domestically and internationally (7, 8). As age increases, immune function declines, and elderly patients often have multiple underlying diseases. Frequent antibiotic use in this population can disrupt gut microbiota, making them more susceptible to CRE infections (9).
The department with the highest CRE isolation rate was the ICU, which aligns with trends observed in most hospitals (10), followed by geriatric and rehabilitation departments. ICU patients often have multiple comorbidities and severe conditions, requiring invasive medical procedures (e.g., tracheostomy, catheter placement) and the use of broad-spectrum antibiotics. Additionally, their weakened immune systems make them highly vulnerable to CRE colonization and transmission (11).
In the geriatric department, most patients are elderly with severe underlying conditions, often receiving prolonged antibiotic treatments and sometimes transferring to ICU wards. The rehabilitation department, a newly established unit specializing in traditional Chinese medicine and functional recovery, also saw a high rate of CRE detection. Many patients in this department are bedridden with pressure ulcers and require invasive procedures such as urethral catheterization, increasing the likelihood of CRE isolation. The relatively high CRE detection rate in non-ICU wards highlights the need to strengthen infection prevention and control measures beyond ICU settings.
Regarding specimen sources, the majority of CRE strains were isolated from sputum and clean midstream urine samples, accounting for 43.85% and 32.09% of cases, respectively. This may be due to the fact that CRE infections predominantly affect open systems, such as the respiratory and urinary tracts, which are more prone to infection when the body's resistance is compromised (12).
The prevalence of CRE isolates from sputum and clean midstream urine specimens in this study may be due to the ease of collecting these specimens and the large volume of samples submitted for testing. This contrasts slightly with other hospitals (13), where CRE isolates are mainly sourced from sputum, followed by purulent secretions and drainage fluids. The isolation of CRE strains from sterile sites holds greater clinical significance. However, the hospital in this study had fewer strains isolated from sterile sites (such as blood, serous fluid, or venous catheters), which warrants closer attention. Further analysis of CRE composition revealed that the predominant CRE bacteria in the hospital were K. pneumoniae, E. coli, and Enterobacter cloacae, which are resistant to carbapenem antibiotics. This aligns with both domestic and international trends (14, 15).
The drug sensitivity results indicated that CRE is resistant to most antibiotics, with susceptibility to only a few, such as tigecycline. This highlights the severe resistance situation of CRE strains, posing substantial limitations on clinical treatment options. Clinical treatment should be guided by drug sensitivity results, with rational selection and combination of antibiotics for the management of CRE infections (16, 17).
The MLST plays a pivotal role in CRE research by analyzing multiple conserved gene fragments to classify strains and identify their evolutionary relationships and transmission patterns. Multilocus sequence typing relies on nucleotide sequencing of housekeeping genes to accurately detect genetic variations in bacteria. This method enables the study of genetic evolution across regions and aids in epidemiological investigations. By comparing laboratory data from different areas, MLST helps determine whether pathogens are related, if they originate from specific clones, and if outbreaks have occurred in hospitals. It can also track CRE transmission routes, identify the source of infection, and, when combined with whole-genome sequencing, conduct in-depth studies on drug resistance mechanisms. The global MLST database supports the monitoring of CRE strains at both local and international levels, assisting public health departments in formulating infection control policies to prevent the spread of CRE.
Carbapenem-resistant Klebsiella pneumoniae is a priority pathogen identified by the WHO as a severe threat to human health, primarily linked to hospital-acquired infections (18, 19). In this study, CRKP accounted for 80.21% of the CRE isolates, with MLST revealing two sequence types, predominantly ST11 (90.91%). This finding is consistent with the predominant genotype and clone type of CRKP in China (20, 21). In China, ST11 CRKP is the leading strain (22), known as the most widespread multidrug-resistant lineage in Asia (23), and was first identified as a hypervirulent strain in China (24).
The detection of the ST15 type is less frequent compared to the ST11 type, but it remains one of the more commonly found strains in China (25). Research has shown (26) that the ST15 CRKP is a high-risk clonal strain that has emerged recently and often leads to hospital outbreaks. ST15, which carries plasmids containing both virulence and resistance genes, has been identified. The most widely prevalent CRKP multilocus sequence types are ST258 and ST11 (22). In the United States and Europe, ST258 CRKP is the dominant type, often associated with localized infections and high mortality rates (27-29). In contrast, ST11-type CRKP is most common in Asia, especially in China (27, 30). The limited diversity of ST types observed in this study could be due to the small number of strains analyzed through MLST typing.
5.1. Conclusions
The continuous emergence of CRE presents significant challenges to clinical treatment and poses a severe threat to patient outcomes. Preventing the occurrence or outbreak of CRE infections in hospitals is crucial. Managing hospital-acquired CRE infections is a complex and systematic endeavor; any oversight or insufficient isolation measures can lead to the spread or even outbreaks of these infections. Effective strategies for preventing and controlling CRE include antibacterial drug management, patient identification and management, as well as environmental and materials management. Implementing these measures in a comprehensive and detailed manner can help reduce the incidence of CRE infections and further decrease mortality rates (31).
Analyzing the epidemiological characteristics and MLST subtypes of hospital CRE-infected patients can effectively aid clinical personnel in understanding the transmission patterns of CRE. This knowledge encourages the strict adherence to antibiotic usage guidelines, including the rational use of carbapenems, and ensures better patient safety.