1. Background
Enteropathogenic Escherichia coli (EPEC), a gram-negative bacterium and opportunistic pathogen, is one of the most common members of the Enterobacteriaceae family. It causes various types of childhood gastrointestinal infections in developing countries (1-3). Often, EPEC-caused diarrhea is watery with abdominal cramps in children. Apart from the frequency of EPEC strains, these strains infect approximately 79,000 children worldwide every year. The infecting potential of EPEC, which is related to its adhesion factors, is usually less than that of other bacteria causing gastrointestinal infections, although the frequency of EPEC is being reported more than other pathogens recently (4-6). Most studies have shown that the pathogenesis of EPEC strains is due to the formation of attaching and effacing lesions in host intestinal cells (7). These lesions can cause physiological changes in intestinal cells. Some of the virulence factors of EPEC strains involved in pathogenesis, such as E. coli attaching and effacing (eae gene) and bundle-forming pilus (bfp gene), were detected (8). These factors cause attachment and invasion of the strains into intestinal cells. The eae and bfp genes are located on the enterocyte effacement pathogenicity island (PAI), locus of enterocyte effacement (LEE), and a plasmid called EPEC adherence factor (EAF), respectively (9, 10).
Enteropathogenic E. coli strains, based on the presence of the EAF plasmid, are characterized into two major groups: (1) Typical enteropathogenic Escherichia coli (tEPEC); and (2) atypical enteropathogenic E. coli (aEPEC) strains (11-13). While tEPEC strains have both eae and bfp genes (eae+ and bfp+), aEPEC strains do not have the bfp gene (eae+ and bfp-). Many studies have shown that aEPEC strains are more frequent than tEPEC strains in developing countries (10). The virulence gene encoding Shiga-like toxin (stx gene) causes bloody diarrhea by enterohemorrhagic E. coli (EHEC) strains. Studies have shown that the existence of eae without either of the stx alleles is a putative marker to confirm EPEC strains. The high prevalence of aEPEC strains has become a serious health issue among children.
2. Objectives
Our purpose was to study the prevalence of tEPEC and aEPEC strains and their antimicrobial resistance among non-diarrheal children under 10 years of age, isolated from schools and welfare centers in Tehran. The grouping of the children was performed according to sex and age groups.
3. Methods
3.1. Specimen Collection
A total of 350 non-diarrheal fecal specimens were collected from children under 10 years of age in schools and welfare centers of Tehran from October 2019 to March 2020. Certain criteria, including the existence of bacterial diarrheal disease (either bloody or watery diarrhea) in individuals for one month prior, individuals above 10 years of age, and antibiotic consumption within one week prior, were excluded from the study. These children were evaluated based on the history of diarrhea period, antibiotic use, and underlying disease. All of the children were categorized into three age groups: < 1 year, 1 to 5 years, and 5 to 9 years. Fecal specimens were collected in sterile containers and transferred to the laboratory for bacteriology experiments (14).
3.2. Escherichia coli Isolation
Initially, specimens were cultured on EMB agar and incubated at 37°C for 24 hours. Isolated colonies with a green metallic sheen and dark purple color were selected as E. coli. Phenotypic confirmation of E. coli isolates was done using the IMViC test, including SIM, TSI, MR-VP, and Simmons citrate agar as biochemical tests. To distinguish commensal E. coli strains from non-commensal strains, they were studied using a molecular test for eae and bfpA genes.
3.3. Genotypic Screening of Enteropathogenic Escherichia coli Strains
Escherichia coli isolates were investigated for the presence of the eae gene. As shown in Table 1, we used a pair of eae primers. The amplification of eae was described as follows: 94°C for 4 minutes for initial denaturation, followed by 30 cycles of 94°C for 33 seconds for denaturation, 64°C for 37 seconds for annealing, 72°C for 40 seconds for extension, and 72°C for 4 minutes as final extension. Grouping of the EPEC strains was done via amplification of bfpA among eae+ strains. The bfpA forward primer (5-AATGGTGCTTGCGCTTGCTGC-3) and reverse primer (5-GCCGCTTTATCCAACCTGGTA-3) were used (15) and amplified as follows: 95°C for 4 minutes for initial denaturation, followed by 34 cycles of 94°C for 47 seconds for denaturation, 54°C for 38 seconds for annealing, 72°C for 55 seconds for extension, and 72°C for 6 minutes as final extension.
The control sample for both eae and bfpA genes was E. coli ATCC 2348/9. To ensure EPEC strains among eae positive strains, both stx-1 and stx-2 primers were used. Amplification of stx-1 and stx-2 was respectively performed as follows: 94°C for 9 minutes for initial denaturation, followed by 31 cycles of 94°C for 31 seconds for denaturation, 59°C for 32 seconds for annealing, 72°C for 30 seconds for extension, and 72°C for 3 minutes as final extension; 95°C for 6 minutes for initial denaturation, followed by 95°C for 31 seconds for denaturation, 63°C for 42 seconds for annealing, 72°C for 40 seconds for extension by 30 cycles, and 72°C for 7 minutes as final extension. Escherichia coli strain EDL 933 was selected as the positive control for both stx-1 and stx-2 genes. All PCR products were analyzed by electrophoresis with 1% agarose gel.
| Primer | Sequence | Fragment Size (bp) | Reference |
|---|---|---|---|
| eae | 229 | (16) | |
| F | 5- TGATAAGCTGCAGTCGAATCC-3 | ||
| R | 5- CTGAACCAGATCGTAACGGC-3 | ||
| bfpA | 326 | (15) | |
| F | 5-AATGGTGCTTGCGCTTGCTGC-3 | ||
| R | 5-GCCGCTTTATCCAACCTGGTA-3 | ||
| stx-1 | 109 | (17) | |
| F | 5-GTGGCATTAATACTGAATTGTCATCA-3 | ||
| R | 5-GCGTAATCCCACGGACTCTTC-3 | ||
| stx-2 | 255 | (18) | |
| F | 5-GGCACTGTCTGAAACTGCTCC-3 | ||
| R | 5-TCGCCAGTTATCTGACATTCTG-3 |
3.4. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed by the disk diffusion method according to the clinical laboratory standards institute (CLSI) guidelines (19). The antimicrobial disks used in the study included meropenem (10 µg), ceftazidime (30 µg), cefotaxime (30 µg), fosfomycin (200 µg), co-trimoxazole (1/25 - 23/75 µg), tetracycline (30 µg), imipenem (10 µg), ciprofloxacin (5 µg), aztreonam (30 µg), amoxicillin-clavulanic acid (20/10 µg), and gentamicin (10 µg). Escherichia coli ATCC 25922 was used as the quality control. As described by the CLSI guidelines (19), all of the isolates were divided into three groups: (1) Sensitive, (2) intermediate resistance, and (3) resistant.
3.5. Statistical Analysis
Data from this study were statistically analyzed using SPSS software version 24. Chi-square and Phi and Cramer's V tests were performed, and P < 0.05 was statistically considered significant.
4. Results
4.1. Specimen Collection
Among the 350 fecal specimens collected, 160 specimens (45.71%) were from boys and 190 specimens (54.29%) from girls. The results showed that 45.71% of specimens were taken from children above 5 years of age, 37.42% were from children between 1 to 5 years of age, and 16.85% were from children under 1 year of age.
4.2. Enteropathogenic Escherichia coli Strains Screening
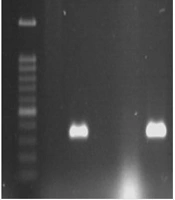
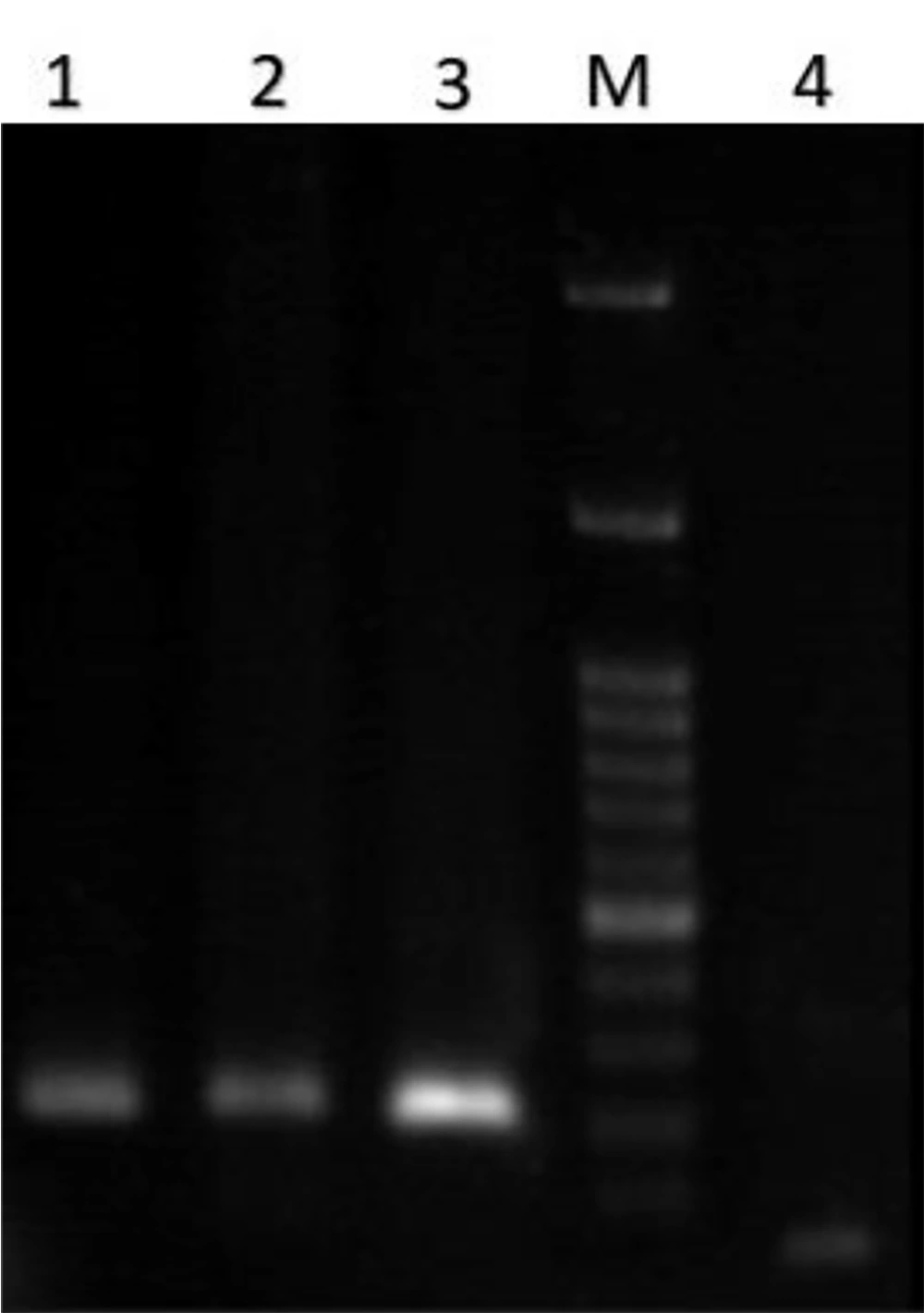
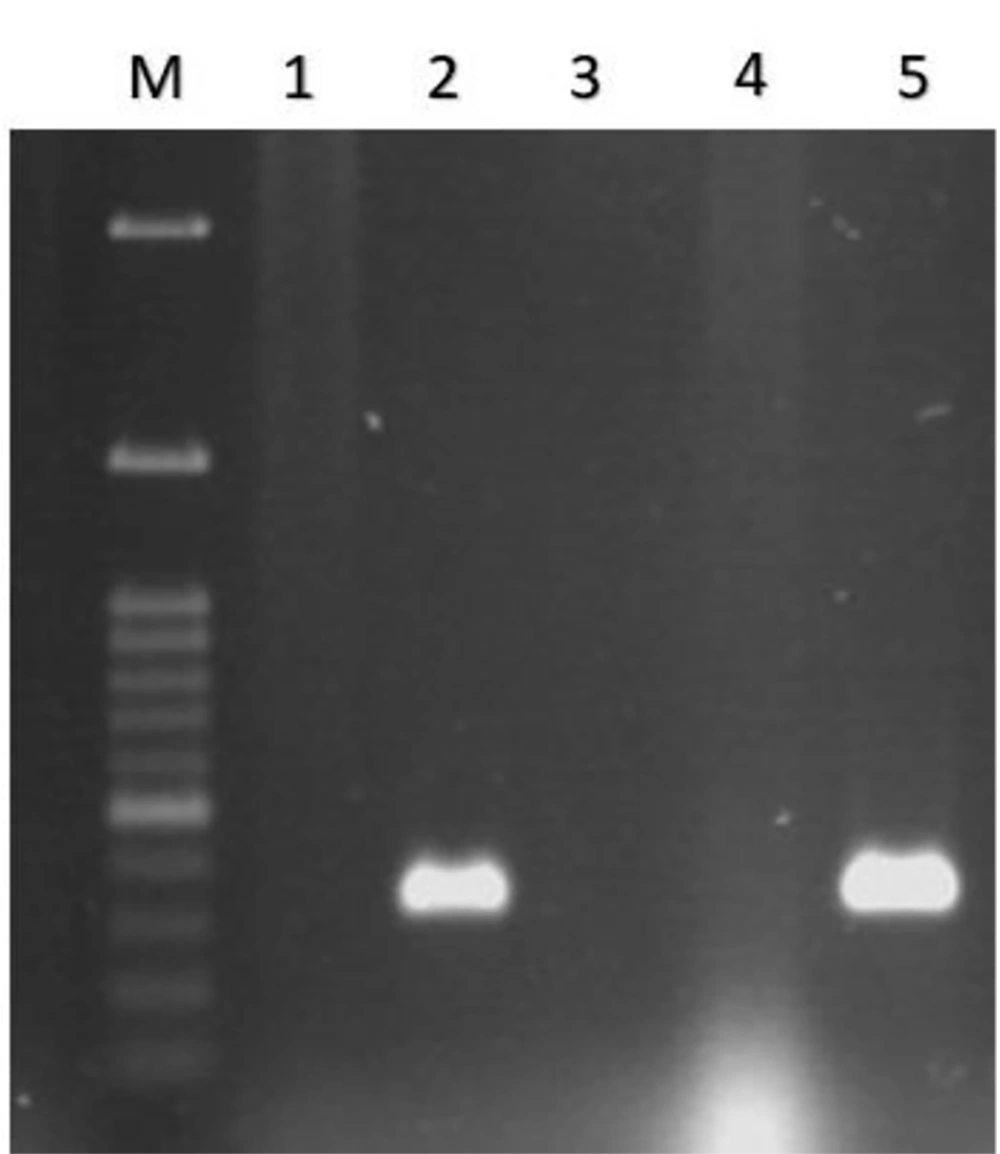
All 350 fecal samples, whose biochemical characteristics were evaluated, were described as E. coli. The next step was to identify EPEC strains using a PCR-based assay. The results of PCR for eae, bfpA, stx-1, and stx-2 genes showed 32% of isolates were eae positive (see Figure 1), and other genes were negative. On the other hand, among the 142 eae-positive strains, 89.43% were bfpA-negative strains and only 1.4% were bfpA positive (see Figure 2), which were typical enteropathogenic E. coli and atypical enteropathogenic E. coli strains, respectively (Table 2).
| Virulence Genes | eae | bfpA | stx-1 | stx-2 | P-Value b | CI c | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | |||
| Age (y) | 0.542 | 0.534 - 0.550 | ||||||||
| < 1 | 16 (14.28) | 43 (12.28) | 0 (0) | 59 (54.12) | 1 (10) | 58 (17.05) | 0 (0) | 59 (16.95) | ||
| 1 - 5 | 43 (38.39) | 88 (25.14) | 3 (100) | 53 (48.62) | 3 (30) | 128 (37.64) | 2(100) | 129 (37.06) | ||
| 5 - 9 | 53 (15.14) | 107 (30.57) | 0 (0) | 67 (61.46) | 6 (60) | 154 (45.29) | 0 (0) | 160 (45.97) | ||
| Total | 112 | 238 | 3 | 109 | 10 | 340 | 2 | 348 | ||
a Values are presented as No. (%) unless otherwise indicated.
b P < 0.05 is significant.
c Confidence interval was computed to 95%.
4.3. Antimicrobial Susceptibility Testing
The study on antimicrobial resistance of aEPEC strains was performed based on the sex and age group of children (see Table 3). Among the antibiotics studied, amoxicillin-clavulanate-resistant E. coli isolates were significantly reported (P-value = 0.03). The highest resistance rate to tetracycline was reported (25% in 5 to 9 years, 28.75% in 1 to 5 years, and 22.22% in < 1-year-old children). On the other hand, the least resistance rate was observed for imipenem (0% in both boys and girls), meropenem (0% in both boys and girls), azithromycin (5% in boys and 2% in girls), and ciprofloxacin (2.38% in boys and 2% in girls).
| Variables | Age (Boy) a | Age (Girl) a | P-Value b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| < 1 | 1 - 5 | 5 - 9 | < 1 | 1 - 5 | 5 - 9 | Age (Boy) | CI c | Age (Girl) | CI c | |
| SXT | 0.735 | 0.727 - 0.744 | 0.585 | 0.575 - 0.594 | ||||||
| S | 0 | 2.38 | 2.38 | 0 | 1.21 | 2.54 | ||||
| I | 0 | 0 | 0 | 0 | 0 | 0.84 | ||||
| R | 22.22 | 25 | 22.61 | 21.95 | 21.95 | 22.88 | ||||
| AMC | 0.038 | 0.034 - 0.042 | 0.030 | 0.026 - 0.033 | ||||||
| S | 3.70 | 0 | 0 | 2.43 | 0 | 0 | ||||
| I | 7.40 | 1.19 | 0 | 2.43 | 1.21 | 5.93 | ||||
| R | 22.22 | 26.19 | 25 | 24.3 | 23.17 | 21.18 | ||||
| AZT | 0.821 | 0.813 - 0.828 | 1.0 | 1.0 | ||||||
| S | 29.62 | 23.80 | 21.42 | 21.95 | 23.17 | 26.17 | ||||
| I | 0 | 1.19 | 2.38 | 0 | 0 | 0 | ||||
| R | 0 | 2.38 | 2.38 | 0 | 1.21 | 0.84 | ||||
| CAZ | 0.77 | 0.768 - 0.784 | 0.74 | 0.737 - 0.754 | ||||||
| S | 18.51 | 20.23 | 20.23 | 21.95 | 23.17 | 23.72 | ||||
| I | 0 | 2.38 | 2.38 | 0 | 0 | 0.84 | ||||
| R | 7.40 | 6 | 2.38 | 0 | 1.21 | 0.84 | ||||
| CP | 0.462 | 0.452 - 0.472 | 1 | 1 | ||||||
| S | 18.51 | 27.3 | 21.42 | 21.95 | 23.17 | 26.27 | ||||
| I | 3.70 | 0 | 2.38 | 0 | 0 | 0 | ||||
| R | 0 | 1.19 | 1.19 | 0 | 1.21 | 0.84 | ||||
| CTX | 0.551 | 0.541 - 0.560 | 1.0 | 1.0 | ||||||
| S | 29.62 | 23.80 | 21.42 | 21.95 | 23.17 | 25.42 | ||||
| I | 3.70 | 2.38 | 0 | 0 | 0 | 0 | ||||
| R | 3.70 | 2.38 | 3.57 | 0 | 1.21 | 0.84 | ||||
| FOS | 0.227 | 0.219 - 0.236 | 0.406 | 0.396 - 0.416 | ||||||
| S | 3.70 | 1.19 | 3.57 | 0 | 2.43 | 2.54 | ||||
| I | 14.81 | 22.61 | 20.23 | 21.95 | 20.17 | 21.18 | ||||
| R | 7.40 | 4.76 | 1.19 | 2.43 | 0 | 2.54 | ||||
| GM | 0.448 | 0.438 | 0.740 | 0.731 - 0.748 | ||||||
| S | 22.22 | 20.23 | 20.23 | 21.95 | 20.17 | 22.03 | ||||
| I | 0 | 6 | 2.38 | 2.43 | 2.43 | 5.08 | ||||
| R | 0 | 2.38 | 2.3 | 0 | 0 | 0 | ||||
| IPM | 0.607 | 0.598 - 0.617 | - | - | ||||||
| S | 22.22 | 26.19 | 25 | 21.95 | 25 | 25.42 | ||||
| I | 0 | 2.38 | 0 | 0 | 0 | 0 | ||||
| R | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| MEM | 0.098 | 0.092 - 0.104 | 0.156 | 0.148 - 0.163 | ||||||
| S | 18.51 | 23.80 | 25 | 21.95 | 25 | 25.42 | ||||
| I | 3.70 | 4.76 | 0 | 2.43 | 0 | 0 | ||||
| R | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| TE | 0.125 | 0.119 - 0.132 | 0.434 | 0.424 - 0.443 | ||||||
| S | 3.70 | 0 | 0 | 0 | 1.21 | 0 | ||||
| I | 0 | 0 | 0 | 0 | 0 | 0.84 | ||||
| R | 22.22 | 28.57 | 25 | 21.95 | 23.17 | 25.42 | ||||
Abbreviations: SXT, trimethoprim-sulfamethoxazole; AMC, amoxicillin-clavulanate; AZT, azithromycin; CAZ, ceftazidime; CTX, cefotaxime; FOS, Fosfomycin, gentamicin; IPM, imipenem; MEM, meropenem; TE, tetracycline; S, sensitive; I, intermediate; R, resistant.
a Values are presented as %.
b P < 0.05 is significant.
c Confidence interval was computed to 95%.
5. Discussion
Enteropathogenic E. coli is the second most frequent pathotype causing traveler's diarrhea in most developing countries (20). Enteropathogenic E. coli strains are not detected with routine protocols of stool culture in clinical laboratories; however, with the use of molecular techniques, EPEC strains have been identified recently (21). Almost all enteropathogenic E. coli strains cause symptomatic diarrhea and have become an important public health issue for children (22). Despite the decrease in the incidence of gastrointestinal infections caused by these strains in developed countries, EPEC strains are increasing in developing countries. On the other hand, the high rate of antibiotic resistance in these strains has generally become a major therapeutic challenge (23). Additionally, there is an increasing health issue in developing communities. Recently, several cases of EPEC colonization in asymptomatic children have been observed (24).
Asymptomatic colonization could increase the prevalence of the strains and even antimicrobial-resistant strains. In the current study, the frequency of aEPEC strains was evaluated among non-diarrheal children. The results of the study showed that the prevalence of asymptomatic colonization of EPEC among children under 10 years was higher. While Chellapandi et al. reported 12 of 50 EPEC strains in children (16), the results of Darbandi et al. (2012) showed that from 158 E. coli isolates, 50 strains were isolated (25). Asymptomatic colonization is much more common in children, with our study showing the most frequent age group for EPEC colonization was reported between 4 to 9 years. In several similar studies, these strains were more frequently reported in ≤ 5-year-old children (26-28). According to the results, prevalence in these age groups is more reported due to the presence of children in public places such as schools and also weak health surveillance in communities (29).
Female patients are much more prevalent than males. Afset et al. reported that the prevalence of aEPEC among female carriers (40.6%) is higher than in males (10). Similarly, our data indicated that girls showed more prevalence than boys. Based on studies, all EPEC strains have the eae gene, and EPEC strains without the bfpA gene are considered aEPEC strains. We differentially studied the frequency of each of the tEPEC and aEPEC strains. Atypical strains are more frequent than typical strains in children. Atypical strains have become important gastrointestinal E. coli pathotypes, with their frequency increasing. Snehaa et al. found 2.5% typical isolates and 7% atypical isolates among 19 EPEC (30). Similarly, our findings demonstrated that these pathotypes are more frequent than typical strains. The results of the study on eae-positive strains showed that most strains did not have the bfpA gene, and only in 3 of the eae-positive strains was the bfpA gene detected. Darbandi et al. similarly reported the bfpA gene (0.7%) among EPEC strains (25).
The higher frequency of aEPEC strains is due to the fact that these strains usually cause non-diarrheal disease, and asymptomatic carriers can easily spread them in the community, leading to a higher frequency compared to tEPEC. In recent years, extensive antibiotic consumption has produced antibiotic-resistant E. coli isolates. Canizalez-Roman et al. showed that EPEC is one of the most prevalent antibiotic-resistant pathotypes (31).
Evaluation of the antibiotic resistance rate in our study exhibited that EPEC strains had higher resistance to amoxicillin-clavulanate, tetracycline, and trimethoprim-sulfamethoxazole than other antibiotics, while among the antibiotics studied, imipenem and gentamicin were reported to have the highest effectiveness on strains. The present study was epidemiologically performed over a limited period and did not study several factors such as the expression of adhesion factors and resistance genes, i.e., beta-lactamase genes. The limitation of this study was that it did not perform more comprehensive antimicrobial resistance tests, both genotypical and phenotypical, and finally did not identify multi-drug resistant strains such as ESBLs and carbapenem-resistant strains, because the aim was to study the epidemiological prevalence of non-diarrheal children infected with enteropathogenic E. coli. Therefore, we recommend that more factors should be investigated.
5.1. Conclusions
The results of our study indicate that asymptomatic carriers of EPEC strains, as well as their antimicrobial resistance rates, are increasing in schools. This is a dangerous alarm because these carriers spread the antimicrobial-resistant strains in the community and can shift the microbial flora pattern.