1. Background
Klebsiella pneumoniae, classified among the ESKAPE pathogens—alongside Enterococcus faecium, Staphylococcus aureus, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.—is recognized for its role in a wide spectrum of human infectious diseases, including abdominal, respiratory, and bloodstream infections, frequently culminating in severe morbidity and mortality (1, 2). Given their remarkable effectiveness in treating K. pneumoniae infections, carbapenems have been designated as the ultimate recourse in combating a multitude of multidrug-resistant bacterial infections. Nonetheless, the injudicious administration of carbapenems has precipitated the emergence of carbapenem-resistant K. pneumoniae (CRKP), profoundly complicating the landscape of clinical anti-infective therapies and eliciting mounting global concern (3). Carbapenemase production serves as the predominant mechanism underlying drug resistance. These carbapenemases are classified into three categories according to the Ambler classification: Class A (blaKPC, blaGES, blaIMI, blaNMC, blaSME), class B (blaIMP, blaVIM, blaNDM, blaGIM, blaSIM, blaSPM), and class D (blaOXA-48).
In recent years, the World Health Organization (WHO) has designated carbapenem-resistant Enterobacteriaceae (CRE) as a priority pathogen, necessitating investment in novel drug development. Specific antibiotics, such as polymyxin, tigecycline, fosfomycin, and aminoglycosides, demonstrate efficacy against a broad range of carbapenemases (4). Furthermore, it is important to note that these antibiotics can evoke more pronounced adverse effects. Notably, approximately 40 - 60% of adult polymyxin users experience nephrotoxicity, and roughly 13% of K. pneumoniae-infected patients exhibit heterogeneous resistance to this antibiotic. Vigilant renal function monitoring is imperative for elderly individuals and those with impaired kidney function.
Despite its effectiveness in treating complex skin and abdominal infections, tigecycline can trigger significant adverse reactions such as coagulation dysfunction and hepatotoxicity. Due to their limited efficacy and suboptimal pharmacokinetic profiles at specific infection sites, the clinical applicability of these agents is restricted. Hence, it is crucial to ascertain robust carbapenemase production before initiating treatment regimens involving high doses and prolonged durations or considering the adoption of combined therapeutic strategies. Failure to do so not only exposes patients to unnecessary adverse effects but also promotes the proliferation of drug-resistant strains.
To address the challenges posed by carbapenemase-producing Enterobacteriaceae, innovative combinations of β-lactams and β-lactamase inhibitors have emerged. Nonetheless, it is important to recognize that these antimicrobial agents may demonstrate heterogeneous sensitivity profiles across different carbapenemase types. Ceftazidime/avibactam (5) serves as a prime example of this phenomenon. Avibactam, acting as a triethylenediamine enzyme inhibitor, effectively inhibits class A and class C β-lactamases. Remarkably, it also displays noticeable inhibitory activity against class D enzymes, particularly targeting blaOXA-10 and blaOXA-48. However, it exhibits limited efficacy against class B β-lactamases, which are categorized as metalloenzymes. Meropenem/vaborbactam and imipenem/relebactam (6) have demonstrated preferential sensitivity primarily towards class A enzymes.
The combined therapy of cefepime/zidebactam has been reported to be effective against a broad spectrum of Enterobacteriaceae, including CRE, KPC-producing strains, metallo-β-lactamases (MBLs), and blaOXA-48-like carbapenemases. However, its sensitivity may be reduced by approximately 80% in the presence of MBLs (7). Research findings have demonstrated that carbapenemase-producing strains result in a worse prognosis for patients infected with carbapenem-resistant bacteria compared to infections caused by other mechanisms, such as efflux pumps or changes in membrane permeability (8). Moreover, this combination therapy has shown promise in improving outcomes for patients with carbapenem-resistant infections. Consequently, the accurate and prompt detection of carbapenemase is crucial for clinical anti-infective treatment, the prevention of nosocomial infections, and the control of CRKP clinical isolates.
2. Objectives
In this study, a retrospective analysis was conducted to compare six methods for detecting CRKP strains collected from 2016 to 2023, using polymerase chain reaction (PCR) sequencing as the reference standard.
3. Methods
3.1. Bacterial Isolates
This was a retrospective, single-center analysis. All strains were identified using the Vitek-2 automated bacterial identification and antimicrobial susceptibility testing system (bioMérieux, France), with confirmation by the Kirby-Bauer (K-B) disk diffusion method. The CRKP was defined as resistance to one or more of the following antibiotics: Imipenem, meropenem, and ertapenem. A total of 88 strains were screened, with quality control strains including ATCC 25922, ATCC BAA-1705, and ATCC BAA-706. Figure 1 illustrates the flowchart of this study.
3.2. Antimicrobial-Resistance Gene Detection
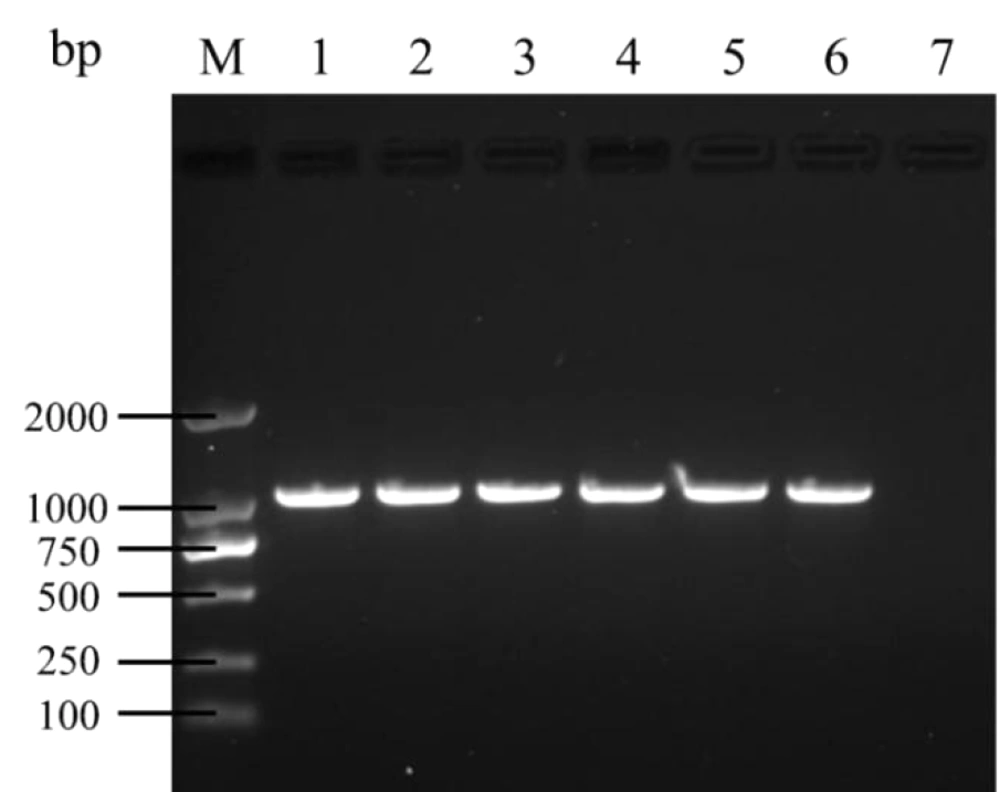
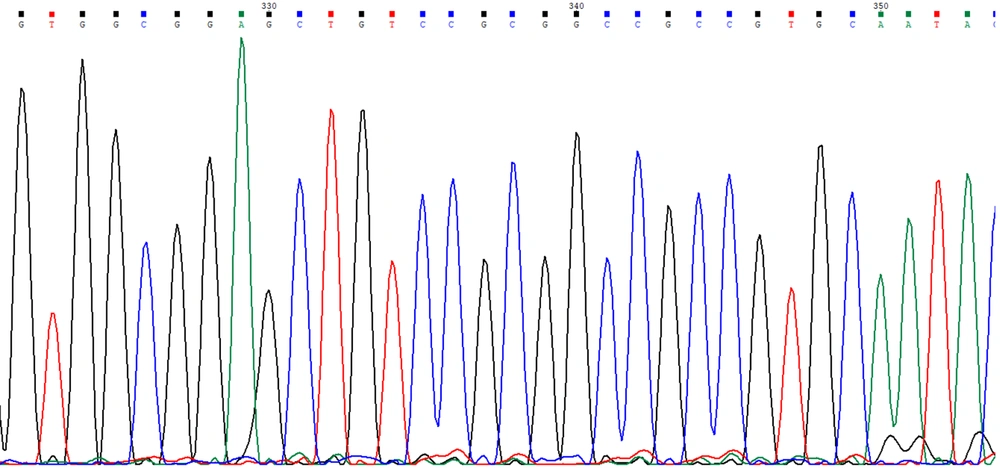
The strains was revived at 35°C for 18 hours, and deoxyribonucleic acid (DNA) extraction was performed using a bacterial genomic DNA extraction kit (Tiangen Biochemical Technology Co., Ltd., Beijing, China). The PCR amplification targeted five carbapenemase genes: Class A (blaKPC), class B (blaNDM, blaVIM, blaIMP), and class D (blaOXA-48). The primers and relevant references are listed in Table 1. After PCR, the products underwent 1.5% agarose gel electrophoresis and were visualized using a gel documentation system. Positive PCR products were selected for sequencing, and the sequence data were aligned using BLAST for analysis.
| Carbapenemase Gene and Primer Sequences | Amplicon Size (bp) | Reference |
|---|---|---|
| blaKPC | 1010 | (9) |
| F: 5’-TGTCACTGTATCGCCGTC-3’ | ||
| R: 5’-CTCAGTGCTCTACAGAAAACC-3’ | ||
| blaNDM | 782 | (10) |
| F: 5’-GCAGCTTGTCGGCCATGCGGGC-3’ | ||
| R: 5’-GGTCGCGAAGCTGCGCACCGCAT-3’ | ||
| blaVIM | 365 | (9) |
| F: 5’-GATGGTGTTTGGTCGCATA-3’ | ||
| R: 5’-CGAATGCGCAGCACCAG-3’ | ||
| blaIMP | 448 | (9) |
| F: 5’-CATGGTTTGGTGGTTCTTG-3’ | ||
| R: 5’-ATAATTTGGCGGACTTTGG-3’ | ||
| blaOXA-48 | 483 | (9) |
| F: 5’-GCGTGGTTAAGGATGAACAC-3’ | ||
| R: 5’-CATCAAGTTCAACCCAACCG-3’ |
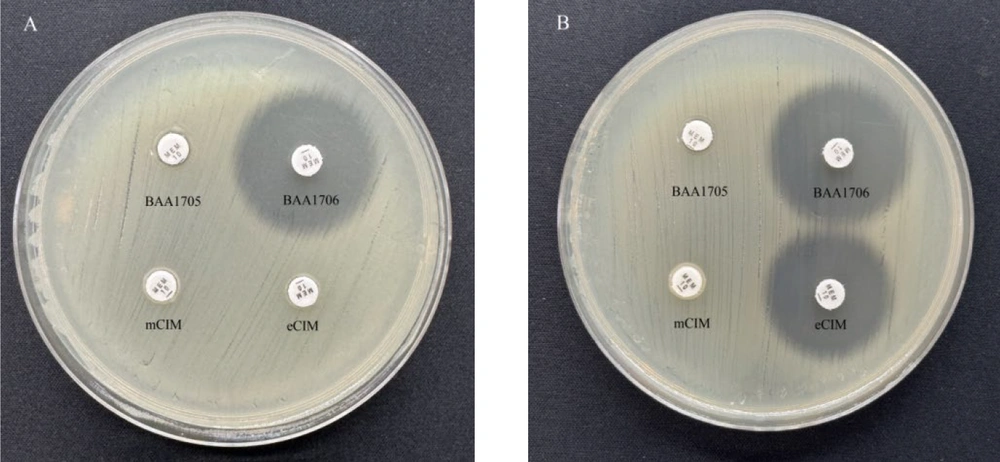
3.3. Modified Carbapenem Inactivation Method and EDTA-Modified Carbapenem Inactivation Method
The operational procedures for modified carbapenem inactivation method (mCIM) and EDTA-modified carbapenem inactivation method (eCIM) adhere to the guidelines outlined in the 2021 CLSI M100 standard. Following overnight incubation, single colonies were meticulously selected and introduced into 2 mL of trypticase soy broth (TSB) using a 1 µL inoculation loop, followed by vortexing for 10 - 15 seconds. Subsequently, a meropenem disk was fully submerged in the bacterial suspension and incubated at 35 ± 2°C for 4 hours. As the incubation of the TSB-meropenem disk suspension neared completion, a 0.5 McFarland turbidity standard suspension of Escherichia coli was prepared using normal saline.
The E. coli suspension was evenly spread onto Mueller-Hinton agar (MHA) plates using a sterile disposable cotton swab. The meropenem disk was then carefully extracted from the TSB using a 10 µL inoculation loop and placed on the inner edge of a test tube, where excess moisture was gently pressed and removed before applying the disk to the pre-inoculated MHA plate with E. coli ATCC 25922. Subsequently, the plate was incubated at 35°C for 18 to 24 hours, after which the diameter of the inhibition zone was measured. For the eCIM assay, a separate clean test tube designated for this purpose was used. Two milliliters of TSB were added, followed by the addition of 20 µL of 0.5 M EDTA solution. The mixture was thoroughly mixed to achieve a final concentration of 5 mM EDTA, after which the subsequent steps were performed identically to those of the mCIM assay.
3.4. Simplified Carbapenem Inactivation Method and Simple EDTA Synergistic Carbapenem Inactivation Method
According to the literature (11), for sCIM, aseptic forceps are used to handle the imipenem disk. One side of the disk is used to collect 3 to 5 overnight colonies from a blood agar plate. The side containing the bacteria is then placed onto an MHA plate inoculated with an E. coli ATCC 25922 suspension, adjusted to a 0.5 McFarland standard. The plate is incubated at 35°C for 16 to 18 hours, followed by measurement of the inhibition zone diameter. Similarly, for esCIM, in accordance with the procedure described in the literature (12), an additional imipenem disk is immersed in a 5 mM EDTA solution in a test tube for 1 minute. Afterward, 3 to 5 overnight colonies are collected and placed onto the same MHA plate. The plate is then incubated at 35°C for 16 to 18 hours, and the diameter of the inhibition zone is measured.
3.5. Cepheid Xpert Carba-R
A single colony is selected from the blood agar plate using a disposable sterile swab. A bacterial suspension with a concentration of 0.5 McFarland is then prepared using physiological saline. Next, 10 μL of the bacterial suspension is aspirated and transferred into a 5 mL processing vial. The vial is securely sealed and vigorously vortexed for approximately 15 seconds using a pipette mixer. Subsequently, 1.7 mL of the mixed sample is carefully introduced into the sample chamber of the test cartridge. The sample is then processed on the GeneXpert platform (Cepheid), with results interpreted after 48 minutes.
3.6. Colloidal Gold Method
The reagent kit (Danna Biotech Co., Ltd., Hunan, China) was retrieved and allowed to equilibrate at room temperature for at least 10 minutes. Subsequently, 300 μL of lysis buffer was transferred into an EP tube, and a single bacterial colony was collected using a 1 μL inoculation loop and added to the lysis buffer. The inoculation loop was gently agitated to release the bacterial sample. After sealing the EP tube, the mixture was thoroughly vortexed for approximately 15 seconds (or longer if the sample was viscous) to ensure adequate bacterial extraction. The tube was then left undisturbed at room temperature for 10 minutes, after which 200 μL of the bacterial suspension was slowly dispensed into the sample well of the test cartridge. The results were observed after 15 minutes.
3.7. Calculation of the Diagnostic Effect
The results of the PCR assay were used as the gold standard. True positive (TP) was defined as cases where both the observation protocol and the gold standard were positive (+). True Negative (TN) referred to cases where both the observation protocol and the gold standard were negative (-). False positive (FP) occurred when the observation protocol was positive (+) while the gold standard was negative (-). False negative (FN) referred to cases where the observation protocol was negative (-) and the gold standard was positive (+). Sensitivity was calculated as TP/(TP + FN) × 100%, and specificity as TN/(TN + FP) × 100%.
3.8. Statistical Analysis
Statistical analysis was performed using SPSS 24.0 software (IBM, USA). The diagnostic performance was evaluated using the kappa test, with values closer to 1 indicating better diagnostic agreement. A P-value of < 0.05 was considered statistically significant.
4. Results
4.1. Results of Carbapenemase Resistance Gene Detection
In this study, a total of 88 CRKP strains were examined, of which 85 tested positive by PCR. The PCR amplification revealed that 81 strains harbored the blaKPC, all carrying the blaKPC-2 genotype. The electropherograms and sequencing chromatograms are shown in Figures 2 and 3. One strain carried the blaNDM-5. Notably, the study also identified the co-occurrence of multiple carbapenemase genes, with two strains harboring both blaKPC-2 and blaNDM-5, and one strain harboring blaKPC-2 and blaIMP-38. This finding highlights the presence of diverse resistance gene profiles under the selective pressure of antimicrobial agents. Additionally, three CRKP strains tested negative by PCR, suggesting the possible presence of alternative resistance mechanisms, such as overexpression of AmpC cephalosporinase or extended-spectrum β-lactamases in conjunction with porin protein deficiency, and modifications in the penicillin-binding protein target sites of carbapenem antibiotics.
4.2. Results of Modified Carbapenem Inactivation Method, EDTA-Modified Carbapenem Inactivation Method, Simplified Carbapenem Inactivation Method and EDTA-Modified Carbapenem Inactivation Method
The results of the mCIM test showed that 80 strains were positive, while the eCIM test identified 5 positive strains. The sCIM test yielded 79 positive strains, and the esCIM test detected 7 positive strains. These findings indicate that serine-producing CRKP is the predominant type in our hospital. The distribution of serine carbapenemases and metallo enzyme is illustrated in Figure 4.
4.3. Results of GeneXpert Carba-R and Colloidal Gold Method
GeneXpert Carba-R detected a total of 83 strains carrying carbapenemase genes, of which 78 strains carried blaKPC, 3 strains carried blaNDM, 1 strain carried blaIMP, and 1 strain carried both blaNDM and blaKPC. Additionally, 3 strains tested negative for carbapenemase genes. The colloidal gold method identified 82 strains carrying carbapenemase genes, with 79 strains carrying blaKPC, 2 strains carrying blaNDM, and 1 strain carrying blaIMP, while 3 strains showed no detectable genes. Some of the test results are presented in Figure 5.
4.4. Comparison of the Performances of Carbapenemase-Detection Methods
The overall sensitivity and specificity values for the six carbapenemase detection tests were as follows: GeneXpert Carba-R demonstrated a sensitivity of 97.65% and a specificity of 100% (kappa = 0.945), while the colloidal gold method showed a sensitivity of 96.47% and a specificity of 100% (kappa = 0.923). The mCIM had a sensitivity of 94.12% and a specificity of 100% (kappa = 0.908), whereas the eCIM exhibited a sensitivity of 67.06% and a specificity of 66.67% (kappa = 0.687). The sCIM showed a sensitivity of 92.94% and a specificity of 100% (kappa = 0.913), while the simple EDTA synergistic carbapenem inactivation method (esCIM) had a sensitivity of 67.06% and a specificity of 100% (kappa = 0.761). The detailed results are provided in Table 2.
| Variables | PCR | Sensitivity (%) | Specificity (%) | Kappa | P-Value | ||
|---|---|---|---|---|---|---|---|
| (+) | (-) | Total | |||||
| GeneXpert Carba-R | 97.65 | 100.00 | 0.945 | < 0.001 | |||
| (+) | 83 | 0 | 83 | ||||
| (-) | 2 | 3 | 5 | ||||
| Total | 85 | 3 | 88 | ||||
| Colloidal gold method | 96.47 | 100.00 | 0.923 | < 0.001 | |||
| (+) | 82 | 0 | 82 | ||||
| (-) | 3 | 3 | 6 | ||||
| Total | 85 | 3 | 88 | ||||
| mCIM | 94.12 | 100.00 | 0.908 | < 0.001 | |||
| (+) | 80 | 0 | 80 | ||||
| (-) | 5 | 3 | 8 | ||||
| Total | 85 | 3 | 88 | ||||
| eCIM | 67.06 | 66.67 | 0.687 | 0.019 | |||
| (+) | 2 | 3 | 5 | ||||
| (-) | 1 | 82 | 83 | ||||
| Total | 3 | 85 | 88 | ||||
| sCIM | 94.94 | 100.00 | 0.913 | < 0.001 | |||
| (+) | 79 | 0 | 79 | ||||
| (-) | 6 | 3 | 9 | ||||
| Total | 85 | 3 | 88 | ||||
| esCIM | 67.06 | 100.00 | 0.761 | 0.002 | |||
| (+) | 2 | 5 | 7 | ||||
| (-) | 1 | 80 | 81 | ||||
| Total | 3 | 85 | 88 | ||||
Abbreviations: mCIM, modified carbapenem inactivation method; eCIM, EDTA-modified carbapenem inactivation method; sCIM, simplified carbapenem inactivation method; esCIM, EDTA synergistic carbapenem inactivation method.
4.5. Characterization of the Six Detection Methods
The Xpert Carba-R test, approved by the US-IVD as an adjunct for infection control, exhibits not only high sensitivity and specificity but also surpasses other test methods by being validated as a diagnostic test directly from blood, eliminating the need for specimen culture. It can rapidly detect carbapenemase genes in rectal swabs to identify colonizing bacteria and can also directly detect these genes in environmental samples for carbapenem-resistant organisms (13, 14). The Xpert Carba-R assay kit, while highly effective, is more expensive and requires specialized instruments, making it less cost-effective for widespread use. Conversely, the colloidal gold method offers an economical alternative and, in this study, demonstrates sensitivity and specificity only marginally lower than that of Xpert Carba-R, rendering it more suitable for use in primary healthcare settings. Table 3 presents a comparison of the primary characteristics of each assay.
| Variables | GeneXpert Carba-R | Colloidal Gold Method | mCIM/eCIM | sCIM/esCIM |
|---|---|---|---|---|
| Classsification of carbapenemase | Detailed classification | Detailed classification | Main categories | Main categories |
| Sample type | Blood cultures, rectal swab, bacterial colonies | Bacterial colonies | Bacterial colonies | Bacterial colonies |
| Operation time before detection | 1 - 2 min | 2 - 3 min | 3 - 5 min | 3 - 5 min |
| Report time | 48 min | 15 - 30 min | 22 - 28 h | 16 - 18 h |
| Interpretation of results | Automatic interpretation | A red stripe appears when there is a quality control line measurement of the diameter of the ring of inhibition | Measurement of the diameter of the ring of inhibition | Subjective if microcolonies present |
| Test reagents and materials | GeneXpert instrument | All supplied by manufacturer | 5mM EDTA, TSB, meropenem disks, MHA plates | 5mM EDTA, imipenem disks, MHA plates |
| Detection of mutated strains | Yes | No | No | No |
| Reagent storing conditions and time | 2 - 28°C, a year and a half | 2 - 30°C, a year | Room temperature, a year | Room temperature, a year |
| Detection principle | Automated real-time quantitative PCR | double antibody sandwich method colloidal gold immunochromatography | Carbapenem hydrolysis assay | Carbapenem hydrolysis assay |
| Approximate cost per test (US$) | 7.00 | 1.50 | < 1.00 | < 1.00 |
Abbreviations: mCIM, modified carbapenem inactivation method; eCIM, EDTA-modified carbapenem inactivation method; sCIM, simplified carbapenem inactivation method.
5. Discussion
The current investigation utilized the mCIM procedure recommended by CLSI, along with eCIM for phenotypic evaluation. The sensitivity and specificity of mCIM in detecting carbapenemase-producing Enterobacteriaceae, as demonstrated in previous studies, typically exceed 93%. In this study, the overall sensitivity and specificity were documented at 94.12% and 100%, respectively, showing strong concordance with PCR-based carbapenemase detection. This finding supports mCIM as a relatively accurate phenotypic detection method. The interpretation of results using the mCIM/eCIM approach is both objective and convenient, facilitated by reliance on defined inhibition zone diameters. However, similar to other phenotypic screening methods, mCIM and eCIM cannot differentiate between serine carbapenemases and metallo-β-lactamases in strains harboring both enzymes. Notably, in this study, a K. pneumoniae strain carrying both blaKPC and blaNDM tested positive with mCIM but negative with eCIM, consistent with previous research findings (15).
Reports on sCIM and esCIM, whose efficacy remains less validated, indicate that these methods operate within a shorter timeframe compared to mCIM and eCIM. Unlike the latter methods, sCIM and esCIM involve scraping three to five colonies using one side of an imipenem disk, omitting the incubation step in TSB before placing the coated side on Mueller-Hinton agar. In this study, the sensitivity and specificity of sCIM and esCIM were 92.94% and 100%, and 66.67% and 94.12%, respectively. These values were slightly lower than those observed with mCIM and eCIM, possibly due to incomplete hydrolysis of imipenem.
The GeneXpert molecular detection platform is an in vitro real-time fluorescence PCR system designed to overcome limitations such as prolonged detection times and reduced sensitivity inherent in traditional bacterial culture and conventional PCR methods. It provides rapid and reliable results and is widely used to detect drug-resistant genes, including those of Mycobacterium tuberculosis, methicillin-resistant S. aureus, and Enterobacteriaceae CRE. The GeneXpert Carba-R system used in this study features an automated analytical framework that integrates sample preparation, nucleic acid extraction, amplification, and real-time PCR detection of target sequences in both simple and complex samples. Developed by Cepheid (USA), the GeneXpert Carba-R system can quickly and accurately detect five common carbapenemase genes, providing a timely basis for clinical decision-making (16).
The GeneXpert system efficiently analyzes detected fluorescence signals and requires no specialized technical training. In this study, it demonstrated an overall sensitivity and specificity of 96.47% and 100%, respectively, consistent with previous reports (17). All identified blaKPC were classified as blaKPC-2. Although no mutated strains were detected, existing data show that GeneXpert Carba-R can identify blaKPC-2 variants in K. pneumoniae, including blaKPC-33, blaKPC-35, blaKPC-71, blaKPC-76, blaKPC-78, and blaKPC-79 (18). Additionally, variants of blaIMP-1, blaIMP-4, and blaIMP-28 were identified (19).
Despite the array of available tools for detecting carbapenemase enzymes, there is currently no single method capable of promptly, accurately, and economically identifying all antibiotic resistance determinants in a rapid and straightforward manner. Clinical microbiology laboratories should consider and select detection methods based on factors such as the reproducibility of test results, ease of operation, cost-effectiveness, simplicity of result interpretation, and rapid detection capabilities. If necessary, optimizing the detection of carbapenem resistance mechanisms may require a combination of phenotypic and genotypic testing methodologies.
This study has certain limitations, including the relatively low detection rate of non-carbapenemase-producing strains, which may lead to an overestimation of specificity. Additionally, the clinical isolates contained only blaKPC, blaNDM, and blaIMP, with blaVIM and blaOXA-48 notably absent. Future research will focus on the collection of additional strains and the expansion of the sample size to facilitate a more comprehensive evaluation of the performance of various carbapenemase detection methods.
5.1. Conclusions
The simplicity and rapid turnaround time of GeneXpert Carba-R support its role in optimizing antimicrobial therapy, thereby enhancing clinical outcomes. Although the colloidal gold method demonstrates slightly lower sensitivity and specificity compared to GeneXpert Carba-R, its cost-effectiveness and minimal equipment requirements make it a practical option for primary healthcare settings.