1. Background
Rapid person-to-person transmission and the increasing number of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) cases have significantly impacted the healthcare system, with millions of confirmed cases and deaths worldwide (1). Severe acute respiratory syndrome coronavirus-2 belongs to the genus Beta coronavirus within the family Coronaviridae. It is a helical, enveloped virus composed of an unsegmented, single-stranded, positive-sense RNA genome of approximately 30 kb, which is capped at the 5'-end and polyadenylated at the 3'-end. Severe acute respiratory syndrome coronavirus-2 primarily transmits through respiratory routes, and the disease it causes is known as Coronavirus disease 2019 (COVID-19) (2). The clinical presentation of COVID-19 can vary widely, ranging from asymptomatic infection to mild disease (81%), or progressing to severe disease (14%) that may result in multi-organ failure and death (5%) (3). Despite extensive research on SARS-CoV-2, the virus's pathophysiology remains incompletely understood and requires further investigation. The pathophysiology of acute respiratory distress syndrome (ARDS) is a multifactorial process, with recent evidence suggesting that host factors, including genetic variations, along with viral and environmental factors, may be involved in the disease’s pathogenesis and outcomes (4-6).
The renin-angiotensin system (RAS) is a hormone system composed of various angiotensin peptides and specific receptors. It is a critical regulator of the cardiovascular, renal, and pulmonary systems and plays a key role in the pathogenesis of many diseases. Severe acute respiratory syndrome coronavirus-2 targets human respiratory cells, including nasal and bronchial epithelial cells and pneumocytes, by binding its viral spike glycoprotein to the ACE2 receptor (7). ACE2 is a 120 kDa single-span transmembrane protein and a zinc-containing metalloenzyme that removes a carboxy-terminal residue from peptide hormones, such as angiotensin II, to produce angiotensin (1-7).
It functions as a negative regulator of the RAS (7-9). From the N-terminal to the C-terminal, ACE2 has four functional and structural domains: The peptidase domain (PD, aa 18 - 615), the collectrin-like domain (CLD, aa 616 - 740), the transmembrane domain (TM, aa 741 - 761), and the intracellular domain (ICD, aa 762 - 805). Additionally, the N-terminal contains a signal peptide (SigP) of 17 amino acids (7-9). Variations in the ACE2 gene can impact the renin-angiotensin pathway by increasing the level of ACE2 in the blood or at specific tissue sites. This can potentially damage the vascular endothelium and lung epithelium, thus increasing susceptibility to viral infections, such as SARS-CoV-2 (10-12).
Several in silico studies have described a correlation between ACE2 polymorphisms and COVID-19 outcomes (13-15). The fourth base of intron 3 in the ACE2 gene contains the rs2285666 polymorphism, also known as G8790A. This polymorphism can alter the process of messenger RNA (mRNA) alternative splicing, which, in turn, affects the level of ACE2 gene expression (13). Recently, researchers have revealed that the ACE2 rs2285666 polymorphism does not influence susceptibility to SARS-CoV-2 infection or the clinical outcome of the disease (16). However, some studies have found an association between ACE2 rs2285666 and COVID-19, suggesting that the GG genotype or the G-allele significantly increases the risk of infection and the severity of COVID-19 (13, 17-19). In contrast, other studies have found no association of this SNP with the disease or its severity (20). The variation in these results may be attributed to differences in ethnicity and the sample populations used in these studies. Therefore, analyzing this polymorphism and its different alleles in a diverse population of various ethnicities in Iran could provide more reliable conclusions.
2. Objectives
The current study aimed to investigate the ACE2 rs2285666 polymorphism in a diverse population of Iranian COVID-19 patients to determine its association with the disease. Additionally, we explored the relationship between the polymorphism and clinical-laboratory data from COVID-19 patients.
3. Methods
3.1. Population of the Study
The clinical samples consisted of 372 subjects who agreed to participate in this multicentric clinical study in Lorestan and Golestan provinces from May to October 2019. We enrolled 201 individuals who developed COVID-19 as contacts. Additionally, a family physician evaluated 172 Iranian medical staff members who showed no clinical signs of COVID-19, and an RT-qPCR test was conducted for confirmation. COVID-19 confirmation was based on the detection of the SARS-CoV-2 genome using RT-qPCR, following World Health Organization (WHO) guidelines. Cases with underlying diseases were excluded from the study population. A total of 5 mL of venous blood was collected from all participants in EDTA-containing tubes for further analysis. Hematological parameters, including erythrocyte sedimentation rate (ESR), hemoglobin (Hb), hematocrit (HCT), platelet (PLT), white blood cell (WBC), red blood cell (RBC), neutrophils, and lymphocytes, were assessed according to the manufacturer’s instructions. In this study, although there was no bias in sample selection, some confounding factors, such as co-morbidities, socioeconomic status, and treatments received, were not considered, which could be limitations of the study.
3.2. DNA Extraction and Genotyping
Genomic DNA was extracted from 350 μL of blood using a DNA isolation kit (Gene All, South Korea). To investigate the presence of the rs2285666 SNP in both groups, a PCR-based restriction fragment length polymorphism (PCR-RFLP) assay was performed. Briefly, 20 μL reaction mixtures were prepared containing 10 μL of Amplicon-PCR Master Mix (10× PCR buffer, 3.0 mM MgCl₂, 0.25 mM dNTPs, 1.5 units of Taq polymerase), 0.5 µmol/L of each primer (F: 5′-CATGTGGTCAAAAGGATATCT-3′, R: 5′-AAAGTAAGGTTGGCAGACAT-3′), 3 µL of purified genomic DNA, and 7 μL of distilled water. Thermal cycling conditions were as follows: One cycle at 95°C for 5 min, followed by 40 cycles consisting of three steps: 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, with a final extension at 72°C for 10 minutes. PCR product digestion was performed by adding 10 U/µL AluI restriction endonuclease (Fermentas, Latvia) (1.5 U), 2 μL of Tango buffer (10×), and 5 μL of PCR products. Distilled water was added to bring the total volume to 20 μL, and the mixture was incubated for 16 hours at 37°C.
3.3. Statistical Analysis
The demographic and clinical data were analyzed using SPSS version 26 software (IBM, Chicago, IL). Hardy-Weinberg equilibrium (HWE) and differences in genotypes were assessed using SNPSTATS online software. Odds ratios (OR) and 95% confidence intervals (CI) were calculated. The chi-square test was applied to determine intergroup differences, while the t-test and Mann-Whitney test were used to compare the means of the two studied groups. A P-value ≤ 0.05 was considered statistically significant for all tests and data analyses.
4. Results
4.1. Genotyping
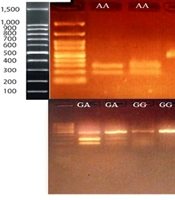
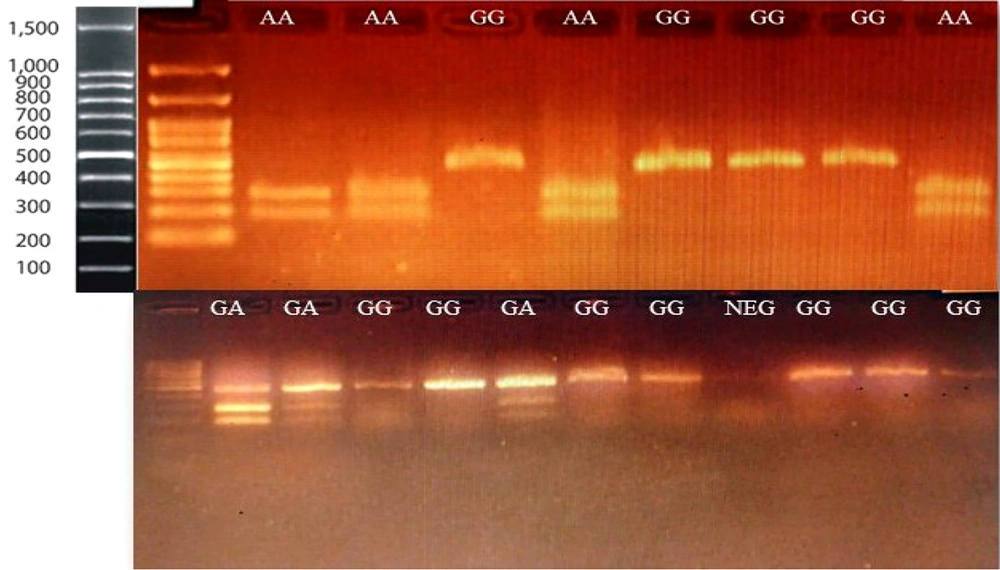
The PCR-RFLP method was used to detect the ACE2 rs2285666 polymorphism. The PCR products were digested using the AluI enzyme. After digestion, the PCR products were electrophoresed on a 2% TBE agarose gel, along with a 100 bp DNA marker, and visualized. A 466 bp fragment was observed in individuals with the GG genotype, three fragments of 185, 281, and 466 bp indicated the GA genotype, and two fragments of 185 and 281 bp corresponded to the AA genotype (Figure 1).
The genotyping of the ACE2 rs2285666 polymorphism was performed using PCR-based restriction fragment length polymorphism (PCR-RFLP). After digestion, fragments of 281 and 185 bp identified the A allele, and a 466 bp band identified the G allele. After electrophoresis, the bands were visualized on Safe-stained agarose gels, and all genotypes were confirmed.
4.2. Characteristics of the Study Population
The present study included 372 participants, of which 201 were COVID-19 confirmed cases, and 171 were negative or non-diagnostic results based on the RT-qPCR test. Among the samples, 173 (46.6%) were women, and 199 (53.4%) were men. The patients’ ages ranged from 0 to 96 years, with an average age of 56 years (Table 1).
| Variables | All Samples | Positive Group (n = 201) | Negative Group (n = 171) |
|---|---|---|---|
| Male | 199 (53) | 94 | 99 |
| Female | 173 (46) | 96 | 77 |
| Mean age (y) | 51 (13) | 56 | 46 |
| Lur | 198 (53) | 100 (50) | 98 (57) |
| Fars | 105 (28) | 76 (38) | 29 (17) |
| Turkmen | 56 (15) | 21 (10) | 35 (21) |
| Balooch | 12 (3) | 4 (2) | 8 (5) |
4.3. Distribution of rs2285666 Genotypes
The genotype frequencies in SARS-CoV-2 positive and negative patients are shown in Table 2. Among females, the frequencies of the GG, GA, and AA genotypes were 61%, 35%, and 4% in the control group and 56%, 29%, and 15% in COVID-19 positive cases, respectively. The frequency of G and A genotypes was 89% and 11% in the control group, and 25% and 75% in the patient group, respectively. The comparison of genotype distributions between patients and the control group across different ethnicities is presented in Table 2. The genotype frequencies differed significantly among Fars participants, as well as across the entire population, for both males (P = 0.029 and P = 0.025, respectively) and females (P = 0.050 and P = 0.050, respectively).
| Ethnicity and Genotypes | Negative Group | Positive Group | P-Value c |
|---|---|---|---|
| Female | |||
| Lur | 0.933 | ||
| GG | 22 ( 56.4) | 30 (55.6) | |
| GA | 15 (38.5) | 22 (40.7) | |
| AA | 2 (5.1) | 2 (3.7) | |
| Fars | 0.050 | ||
| GG | 11 (73.3) | 17 (54.8) | |
| GA | 4 (28.6) | 3 (9.3) | |
| AA | 0 (0.0) | 11 (34.4) | |
| Turkmen | 0.186 | ||
| GG | 11 (68.8) | 7 (63.6) | |
| GA | 5 (31.3) | 2 (18.2) | |
| AA | 0 (0.0) | 2 (18.2) | |
| Balooch | - | ||
| GG | 3 (42.9) | - | |
| GA | 4 (57.1) | ||
| AA | - | ||
| Total | 0.050 | ||
| GG | 47 (61) | 54 (56.2) | |
| GA | 27 (35.1) | 28 (29.2) | |
| AA | 3 (3.9) | 14 (14.6) | |
| Male | |||
| Lur | 0.594 | ||
| G | 50 (84.7) | 37 (80.4) | |
| A | 9 (15.3) | 9 (19.6) | |
| Fars | 0.029 | ||
| G | 23 (100.0) | 25 (69.4) | |
| A | 0 (0.0) | 11 (30.6) | |
| Turkmen | 0.578 | ||
| G | 17 (89.5) | 8 (72.7) | |
| A | 2 (12.5) | 3 (27.3) | |
| Balooch | 0.250 | ||
| G | 4 (100) | 0 (0.0) | |
| A | 0 (0.0) | 1 (100.0) | |
| Total | 0.025 | ||
| G | 94 (89.5) | 70 (74.5) | |
| A | 11 (10.5) | 24 (25.5) |
The allele frequencies in the control group, patients, and total population were 0.83, 0.72, and 0.77 for the G allele, and 0.17, 0.28, and 0.23 for the A allele, respectively. The population was found not to be in HWE in both the positive and negative groups (P ≤ 0.05) (Table 3). Considering that ACE2 is an X-linked gene, females were studied to determine the inheritance pattern of the polymorphism. For both co-dominant (P = 0.046) and recessive (P = 0.014) inheritance types, there was a significantly higher likelihood of COVID-19 infection among individuals with the AA genotype, with OR of OR = 4.06 (1.10 - 15.00) and OR = 4.21 (1.16 - 15.24), respectively (Table 4). Additionally, there was a significantly higher frequency of the A allele in COVID-19 patients (P = 0.007).
| Alleles | Frequency | Hardy-Weinberg (P-Value) | ||||
|---|---|---|---|---|---|---|
| Control | Patient | Total | Control | Patient | Total | |
| G | 0.83 | 0.72 | 0.77 | 1 | 0.0057 | 0.029 |
| A | 0.17 | 0.28 | 0.23 | |||
Allele Frequencies of rs2285666 among Participants a
| Model and Genotypes | SNP Association with Response Group (n = 173, Crude Analysis) | |||||
|---|---|---|---|---|---|---|
| Control Group | Patient Group | OR (95% CI) | P-Value | AIC | BIC | |
| Co-dominant | 0.046 | 237.6 | 247.1 | |||
| G/G | 47 (61) | 54 (56.2) | 1.00 | |||
| G/A | 27 (35.1) | 28 (29.2) | 0.90 (0.47 - 1.74) | |||
| A/A | 3 (3.9) | 14 (14.6) | 4.06 (1.10 - 15.00) | |||
| Dominant | 0.52 | 241.3 | 247.6 | |||
| G/G | 47 (61) | 54 (56.2) | 1.00 | |||
| G/A-A/A | 30 (39) | 42 (43.8) | 1.22 (0.66 - 2.24) | |||
| Recessive | 0.014 | 235.7 | 242 | |||
| G/G-G/A | 74 (96.1) | 82 (85.4) | 1.00 | |||
| A/A | 3 (3.9) | 14 (14.6) | 4.21 (1.16 - 15.24) | |||
| Overdominant | 0.41 | 241.1 | 247.4 | |||
| G/G-A/A | 50 (64.9) | 68 (70.8) | 1.00 | |||
| G/A | 27 (35.1) | 28 (29.2) | 0.76 (0.40 - 1.45) | |||
| Log-additive | 0.13 | 239.4 | 245.7 | |||
| - | - | - | 1.43 (0.90 - 2.27) | |||
4.4. Association of the rs2285666 Polymorphism with Clinical Symptoms and Laboratory variables
The study of clinical symptoms showed that patients exhibited fever (48.2%), cough (42.3%), muscle pain (17.4%), respiratory distress (52.2%), diabetes (12.9%), heart disease (8.4%), and high blood pressure (6.5%). The results showed that female patients with the GG genotype had significantly higher frequencies of fever (P = 0.041), respiratory distress (P = 0.015), and high blood pressure (P = 0.037). There were also significantly higher frequencies of respiratory distress among all female participants (P = 0.003). Among males with the G genotype and all male participants, the patient group showed significantly higher frequencies of fever (P = 0.008 and P = 0.004, respectively) and respiratory distress (P = 0.003 and P < 0.001, respectively) (Table 5).
| Variables | Fever | Cough | Muscle pain | Respiratory Distress | Diabetes | Heart Disease | Blood Pressure | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACE2 G8790A | No | Yes | No | Yes | No | Yes | No | Yes | No | Yes | No | Yes | No | Yes |
| Female | ||||||||||||||
| GG | ||||||||||||||
| Control | 32 (68.1) | 15 (31.9) | 31 (65.9) | 16 (34.1) | 40 (85.1) | 7 (14.9) | 32 (68.1) | 15 (31.9) | 41 (87.2) | 6 (12.8) | 44 (93.6) | 3 (6.4) | 47 (100.0) | 0 (0.0) |
| Case | 26 (48.1) | 28 (51.9) | 32 (59.2) | 22 (40.8) | 42 (77.8) | 12 (22.2) | 24 (44.4) | 30 (55.6) | 46 (85.2) | 8 (14.8) | 48 (88.9) | 6 (11.1) | 48 (88.9) | 6 (11.1) |
| P- value c | 0.041 | 0.532 | 0.316 | 0.015 | 1.000 | 0.511 | 0.037 | |||||||
| GA | ||||||||||||||
| Control | 15 (55.6) | 12 (44.4) | 17 (62.9) | 10 (37.1) | 26 (96.3) | 1 (3.7) | 20 (74.1) | 7 (25.9) | 26 (96.3) | 1 (3.7) | 25 (92.6) | 2 (7.4) | 25 (92.6) | 2 (7.4) |
| Case | 16 (57.1) | 12 (42.9) | 16 (57.1) | 12 (42.9) | 23 (82.1) | 5 (17.9) | 17 (60.7) | 11 (39.3) | 22 (78.6) | 6 (21.4) | 26 (92.9) | 2 (7.1) | 26 (92.9) | 2 (7.1) |
| P- value c | 1.000 | 0.779 | 0.192 | 0.372 | 0.100 | 1.000 | 1.000 | |||||||
| AA | ||||||||||||||
| Control | 2 (66.6) | 1 (43.4) | 2 (66.6) | 1 (43.4) | 1 (43.4) | 2 (66.6) | 1 (43.4) | 2 (66.6) | 3 (100) | 0 (0.0) | 3 (100.0) | 0 (0.0) | 3 (100.0) | 0 (0.0) |
| Case | 5 (35.7) | 9 (64.3) | 8 (57.1) | 6 (42.9) | 12 (85.7) | 2 (14.3) | 5 (35.7) | 9 (64.3) | 11 (78.6) | 3 (21.4) | 13 (92.9) | 1 (17.1) | 13 (92.9) | 1 (17.1) |
| P- value c | 1.000 | 1.000 | 0.331 | 1.000 | 1.000 | 1.000 | 1.000 | |||||||
| Total | ||||||||||||||
| Control | 49 (63.6) | 28 (36.4) | 50 (64.9) | 27 (35.1) | 67 (87.1) | 10 (11.9) | 53 (68.8) | 24 (31.2) | 70 (90.9) | 7 (9.1) | 72 (93.5) | 5 (6.5) | 75 (97.4) | 2 (2.6) |
| Case | 47 (48.9) | 49 (51.1) | 56 (58.3) | 40 (41.7) | 77 (80.2) | 19 (19.8) | 46 (47.9) | 50 (52.1) | 79 (82.3) | 17 (17.7) | 87 (90.6) | 9 (9.4) | 87 (90.6) | 9 (9.4) |
| P- value c | 0.059 | 0.519 | 0.206 | 0.003 | 0.170 | 0.593 | 0.203 | |||||||
| Male | ||||||||||||||
| G | ||||||||||||||
| Control | 67 (71.3) | 27 (28.7) | 62 (65.9) | 32 (44.1) | 81 (86.2) | 13 (13.8) | 63 (67.1) | 31 (32.9) | 93 (98.9) | 1 (1.1) | 86 (91.5) | 8 (8.5) | 88 (93.6) | 6 (6.4) |
| Case | 34 (48.6) | 36 (51.4) | 42 (60) | 28 (40) | 57 (81.4) | 13 (18.6) | 29 (41.4) | 41 (58.6) | 64 (91.4) | 6 (8.6) | 65 (92.9) | 5 (7.1) | 66 (94.3) | 4 (5.7) |
| P- value c | 0.008 | 0.594 | 0.486 | 0.003 | 0.115 | 0.764 | 1.000 | |||||||
| A | ||||||||||||||
| Control | 8 (72.7) | 3 (17.3) | 4 (36.4) | 7 (63.6) | 11 (100.0) | 0 (0.0) | 9 (81.8) | 2 (18.2 | 10 (90.9) | 1 (9.1) | 11 (100.0) | 0 (0.0) | 11 (100.0) | 0 (0.0) |
| Case | 12 (50.0) | 12 (50.0) | 7 (29.2) | 17 (70.8) | 21 (87.5) | 3 (12.5) | 10 (41.7) | 14 (58.3) | 21 (87.5) | 3 (12.5) | 21 (87.5) | 3 (12.5) | 24 (100.0) | 0 (0.0) |
| P- value c | 0.446 | 0.683 | 0.534 | 0.060 | 1.000 | 0.534 | - | |||||||
| Total | ||||||||||||||
| Control | 75 (71.4) | 30 (28.6) | 66 (62.9) | 39 (37.1) | 92 (87.6) | 13 (12.4) | 73 (69.5) | 32 (30.5) | 103 (98.1) | 2 (1.9) | 97 (92.4) | 8 (7.6) | 99 (94.3) | 6 (5.7) |
| Case | 46 (48.9) | 48 (51.1) | 49 (52.1) | 45 (47.9) | 78 (82.9) | 16 (17.1) | 39 (41.5) | 55 (58.5) | 85 (90.4) | 9 (9.6) | 86 (91.5) | 8 (8.5) | 90 (95.7) | 4 (4.3) |
| P-value c | 0.004 | 0.212 | 0.287 | < 0.001 | 0.065 | 1.000 | 1.000 | |||||||
In female patients, there was a significant increase in the ESR and a significant decrease in lymphocyte levels among both GG genotype females (P = 0.02 and P < 0.001, respectively) and all female participants (P = 0.031 and P = 0.006, respectively). In male patients with the A genotype, there was a significant increase in neutrophils (P = 0.007) and a significant decrease in lymphocyte levels (P = 0.004). In male patients with the G genotype, the Hb level was significantly decreased (P = 0.016). Among all male patients, there was a significant increase in ESR (P = 0.008) and neutrophils (P = 0.013), along with a significant decrease in RBC count (P = 0.029), Hb (P = 0.001), and HCT (P = 0.004) (Table 6). It should be noted that the results of this study could be influenced by limitations, such as the small number of individuals with the AA and A genotypes, as well as potential confounding factors that may affect laboratory values (e.g., age, medication, severity of COVID-19).
| ACE2 Polymorphism | ESR | WBC | RBC | Hb | Hematocrit | Platelet | Neutrophil | Lymphocytes |
|---|---|---|---|---|---|---|---|---|
| Female | ||||||||
| GG | ||||||||
| Control | 20.923 (20.2791) | 7.927 (4.8733) | 4.573 (1.1016) | 13.277 (2.9285) | 38.650 (7.9791) | 191.23 (77.563) | 60.43 (23.714) | 28.29 (12.669) |
| Case | 39.385 (23.3685) | 8.865 (4.4673) | 4.177 (.7337) | 12.437 (2.3279) | 36.223 (6.0680) | 203.58 (85.313) | 68.21 (28.567) | 16.77 (10.685) |
| P-value c | 0.020 | 0.447 | 0.146 | 0.220 | 0.199 | 0.576 | 0.292 | < 0.001 |
| GA | ||||||||
| Control | 33.889 (21.7913) | 7.433 (3.7397) | 4.533 (.4850) | 12.913 (2.9895) | 38.853 (4.3536) | 214.40 (83.494) | 53.93 (30.115) | 26.80 (10.605) |
| Case | 44.769 (32.2727) | 6.446 (1.9509) | 4.375 (.5252) | 13.546 (1.2079) | 38.058 (3.9715) | 212.46 (56.971) | 65.33 (19.482) | 25.42 (13.571) |
| P-value c | 0.390 | 0.356 | 0.352 | 0.446 | 0.561 | 0.932 | 0.158 | 0.739 |
| AA | ||||||||
| Control | 33.500 (41.7193) | 7.950 (2.6163) | 3.750 (.7778) | 11.100 (3.8184) | 31.350 (6.7175) | 285.50 (122.329) | 62.00 (15.556) | 25.50 (9.192) |
| Case | 37.000 (27.8388) | 6.120 (2.8700) | 4.38 (.3962) | 13.340 (2.0428) | 38.500 (4.6707) | 154.20 (92.356) | 68.40 (23.681) | 25.60 (21.102) |
| P-value c | 0.915 | 0.473 | 0.190 | 0.333 | 0.158 | 0.174 | 0.703 | 0.993 |
| Total | ||||||||
| Control | 26.833 (22.3270) | 7.738 (4.3031) | 4.515 (.8978) | 13.026 (2.9415) | 38.354 (6.7886) | 204.97 (82.296) | 57.95 (25.780) | 27.55 (11.505) |
| Case | 40.881 (26.1331) | 7.825 (3.8392) | 4.267 (.6412) | 12.888 (2.0333) | 37.089 (5.2866) | 203.09 (77.335) | 67.21 (25.071) | 20.42 (13.137) |
| P-value c | 0.031 | 0.915 | 0.137 | 0.797 | 0.294 | 0.905 | 0.074 | 0.006 |
| Male | ||||||||
| G | ||||||||
| Control | 27.893 (21.1351) | 9.649 (11.8762) | 4.772 (.8821) | 15.026 (2.9615) | 41.681 (7.2753) | 205.70 (83.349) | 66.46 (23.240) | 21.83 (13.124) |
| Case | 37.906 (24.1546) | 7.769 (4.3846) | 4.533 (.6895) | 13.758 (1.8831) | 39.079 (5.2343) | 195.53 (94.043) | 74.09 (13.042) | 20.04 (11.404) |
| P-value c | 0.095 | 0.321 | 0.162 | 0.016 | 0.057 | 0.588 | 0.057 | 0.492 |
| A | ||||||||
| Control | 18.400 (24.2961) | 6.188 (2.1490) | 5.150 (.7597) | 15.013 (2.1735) | 41.725 (4.6879) | 232.88 (50.309) | 55.13 (23.314) | 29.25 (8.763) |
| Case | 52.000 (40.3044) | 9.500 (5.4299) | 4.442 (.7821) | 13.250 (2.2857) | 37.950 (5.6843) | 227.92 (131.524) | 77.58 (8.878) | 16.50 (8.274) |
| P-value c | 0.113 | 0.120 | 0.060 | 0.102 | 0.137 | 0.921 | 0.007 | 0.004 |
| Total | ||||||||
| Control | 25.730 (21.3758) | 9.200 (10.6388) | 4.835 (.8433) | 15.070 (2.7676) | 41.845 (6.7401) | 210.15 (76.883) | 65.44 (22.684) | 22.56 (12.735) |
| Case | 41.511 (29.1194) | 8.636 (5.1439) | 4.514 (.7103) | 13.568 (2.0372) | 38.278 (6.3821) | 206.14 (101.863) | 74.18 (14.051) | 20.10 (12.899) |
| P-value c | 0.008 | 0.710 | 0.029 | 0.001 | 0.004 | 0.808 | 0.013 | 0.295 |
5. Discussion
The clinical outcomes of SARS-CoV-2 infection vary widely among individuals, ranging from mild respiratory symptoms to multi-organ failure, which can lead to mortality (21). ACE2 serves as the functional receptor for viral attachment and penetration into host cells. In females, the frequencies of GG, GA, and AA genotypes were 61%, 35%, and 4% in the control group, and 56%, 29%, and 15% in COVID-19 positive patients, respectively. Among male participants, the prevalence of G and A genotypes was 89% and 11% in the control group, and 25% and 75% in the patient group, respectively. The polymorphism frequencies were not in HWE in both the positive and negative groups (P < 0.05). Logistic regression analysis revealed a significant difference for the AA genotype in both co-dominant and recessive inheritance models, with OR of OR = 4.06 (1.10 - 15.00) and OR = 4.21 (1.16 - 15.24), respectively. This study identified a strong association between the ACE2 rs2285666 AA or A genotypes and the likelihood of contracting COVID-19.
Several studies have shown that different SNPs in the ACE2 gene may contribute to the severity of COVID-19 and other diseases, such as hypertension, type 2 diabetes mellitus, and cardiovascular diseases (7). Numerous clinical studies have investigated these SNPs, including rs4830542, rs4240157, rs4646155, rs233575, rs2074192, rs2285666, and rs2106809 (7, 22, 23). Among these characterized SNPs, the G8790A (rs2285666, G > A) polymorphism is the most well-known and has been associated with susceptibility to COVID-19 and its severity (24). Most studies have been limited to in silico analyses, and there are few reports exploring the relationship between clinical outcomes and the rs2285666 polymorphism (12-16). In a recent study, Karakas Celik et al. analyzed ACE I/D, ACE2 rs2106809, and rs2285666 polymorphisms in 155 hospitalized Turkish COVID-19 patients, who were divided into three groups (mild, moderate, and severe). They found no association between these polymorphisms and COVID-19 severity (16).
In our study, we analyzed samples from 372 Iranian participants for the rs2285666 polymorphism. The results revealed differences in the genotype and allele frequencies between SARS-CoV-2-positive individuals and the control group. In contrast to the findings of Karakas Celik et al., we found a significant association between the ACE2 rs2285666 AA genotype or A-allele and the risk of developing COVID-19. Although a significant association was observed, it should be noted that some limitations, such as the small sample size of AA genotype cases, may affect the study. These findings provide promising perspectives for future research.
Srivastava et al. conducted a haplotype analysis and reported significantly lower rates of infection and case-fatality rate (CFR) for the alternate A allele of rs2285666 among Indian populations (13). Additionally, they found that the frequency of this allele was significantly higher in Indian populations compared to European, American, or African populations. However, while their study provided haplotype analysis, it did not establish a direct clinical association between the severity of COVID-19 and the rs2285666 polymorphism (13). In contrast, a cohort study found that the GG genotype or G allele of rs2285666 was significantly associated with higher infection (two-fold) and mortality (three-fold) rates (18). The current study found a significant association between the rs2285666 AA and A genotypes and COVID-19, along with some clinical-laboratory features, which is consistent with previous studies.
Sienko et al. studied 188 SARS-CoV-2-positive patients in two groups: Mild and severe COVID-19 cases that required hospitalization. The results indicated an increased risk of severity associated with specific ACE2 gene polymorphisms. The highest correlation was found for the variants rs2285666 (AA allele, OR = 2.12, P = 0.0189), rs2074192 (TT allele, OR = 2.05, P = 0.0016), rs4646174 (GG allele, OR = 1.93, P = 0.0016), rs4646156 (TT allele, OR = 1.71, P = 0.008), and rs2158083 (TT allele, OR = 1.84, P = 0.0025). The results of this study are consistent with the present study, which suggests the role of AA and A genotypes in COVID-19 infection susceptibility (25).
Najafi and Mahdavi examined 146 hospitalized COVID-19 patients, including 102 with mild and 44 with severe cases. There was no statistically significant relationship between the studied groups and genotype frequencies (P = 0.8472). Although the ACE2 rs2285666 polymorphism was not associated with disease severity, the SNP was significantly associated with ALT, ESR, and P. The results of this study were not consistent with those of the present study (20).
Alimoradi et al. studied 129 samples, including negative controls, mild, and severe groups. The results indicated an association between the ACE2 rs2285666 genotype (GG) and susceptibility to COVID-19 infection (P = 0.008; OR 5.0, 95% CI: 1.4 - 17.8), but no association with infection severity. These results were not consistent with our findings, which showed an association between the AA and A genotypes with the disease (17). The rs2285666 variant regulates alternative splicing and increases the expression level of the ACE2 receptor gene by up to 50%, potentially playing a significant role in SARS-CoV-2 susceptibility. In a study by Li, serum levels of ACE2 were higher in individuals with the AA genotype compared to those with the GG and GA genotypes (26). The overexpression of ACE2 in individuals with the AA genotype may be inconsistent with the current study’s results, which found the AA genotype to be associated with COVID-19. Since the rs2285666 variant is considered to be a splicing enhancer site, measuring serum ACE2 protein could potentially help predict the progression of COVID-19.
In the current study, we analyzed a large number of samples to explore the role of the ACE2 rs2285666 polymorphism in SARS-CoV-2 viral infection and to determine the prevalence of ACE2 rs2285666 among Iranian populations with different ethnic backgrounds. It is important to note that our results reflect a general population with some limitations in sample selection, rather than being the outcome of a highly selective or restricted study. The exact role of the ACE2 rs2285666 polymorphism in COVID-19 disease outcomes remains controversial. This controversy may be attributed to various confounding factors such as disease status, sample collection methods (including underlying health conditions or exposure levels), demographics (e.g., age, sex), and ethnic factors. The sample sizes for some ethnic groups (especially the Balooch) were small, making it challenging to draw reliable conclusions about genotype distribution.
5.1. Conclusions
Our study provides evidence of a relationship between the ACE2 rs2285666 (G8790A) polymorphism and COVID-19 susceptibility. The data from this study can be extended to general populations of different ethnicities, contributing to the understanding of the pathogenicity of SARS-CoV-2. Our findings highlight the need for large-scale studies to identify variations and polymorphisms in the ACE2 gene and determine their role in the susceptibility and severity of COVID-19 across different genetic backgrounds. Some limitations exist, including confounding factors in sample selection and the low number of participants from certain ethnic groups. Despite these limitations, the promising results of this study could guide future research in this area. The authors recommend further studies involving larger and more diverse populations to investigate the impact of this polymorphism on COVID-19 susceptibility and severity.