1. Background
Coronaviruses, a group of positive-sense single-strand RNA viruses with an envelope and genome sizes ranging from 26 to 32 kilobases, belong to the Coronaviridae family. The recent SARS-CoV-2 outbreak has rapidly spread to numerous countries, causing an acute febrile illness with severe respiratory distress syndrome (1-3). During SARS-CoV-2 infection, infected cells secrete significant amounts of chemokines and cytokines, including IL1, IL6, IL8, IL21, TNF-α, and MCP1, in response to the virus. These chemokines and cytokines recruit lymphocytes and leukocytes to the site of infection (4, 5). Additionally, coronaviruses can infect macrophages, which then present viral antigens to T-cells. This process activates and differentiates T-cells, leading to cytokine production associated with various T-cell subsets (e.g., Th17). The subsequent massive release of cytokines amplifies the immune response. However, sustained production of these mediators due to viral persistence negatively impacts the activation of natural killer (NK) cells and CD8 T-cells.
CD8 T-cells, in contrast, produce potent mediators to clear coronaviruses. However, the dysregulated immune response, particularly the cytokine storm, is one of the primary reasons for poor prognosis in patients with SARS-CoV-2. This evidence highlights the critical role of inflammation in this disease and underscores the potential for developing new therapeutic and diagnostic approaches (6, 7). MicroRNAs are a class of non-coding RNA, approximately 22 nucleotides in length, that regulate gene expression through mRNA degradation or translational repression at the post-transcriptional level, often by targeting the 3' untranslated region (3-UTR) of their target genes. These RNAs play crucial roles in various biological processes, especially in immune and inflammatory responses, as demonstrated in numerous studies (8-10).
miR-26a is one of the key microRNAs involved in the inflammatory response. It has been strongly expressed in the lung tissues of asthmatic mice and in the bronchoalveolar lavage fluid (BALF) of asthmatic children, suggesting its critical role in airway inflammation (11). Furthermore, miR-26a regulates allergic inflammation through a feedback mechanism involving miR-26a/-26b and COX-2 (12). The miR-125 family is one of the most notable miRNA families involved in host immune responses, malignancy, and disease progression (13, 14). miR-125b has been reported to specifically target and inhibit the expression of a gene encoding 5-lipoxygenase, an essential enzyme in the biosynthesis of leukotrienes, which are crucial for innate immune responses and inflammatory processes (15). Additionally, both miR-26a and miR-125b have been shown to inhibit TNF-α expression via different pathways, emphasizing their significant roles in inflammation (9, 10).
2. Objectives
The current study aims to evaluate the expression levels of miR-26a and miR-125b in the WBCs of patients with mild symptoms compared to those in critical condition who are hospitalized in the ICU. This research examines these expression levels within the context of patients with mild symptoms and those with severe conditions requiring intensive care.
3. Methods
3.1. Participants and Methods for Clinical Sampling
Applying the inclusion and exclusion criteria (none of the participants had any other known underlying diseases such as diabetes, autoimmune diseases, or immunodeficiencies), patients were separated into two groups. The first group consisted of patients with a poor prognosis, presenting with severe symptoms (such as cough, fatigue, and fever), low oxygen levels (below 89%), and chronic illnesses (including obesity, hypertension, chronic lung disease, or diabetes). The second group included patients with a good prognosis, exhibiting mild symptoms (such as cough, fatigue, and fever), oxygen levels of 89% or higher, and no chronic illnesses (including obesity, hypertension, chronic lung disease, or diabetes). These classifications were made after admission and hospitalization during the first 10 days. All patients were diagnosed using real-time PCR and were hospitalized at the hospital between January 2021 and March 2021. Table 1 provides detailed clinicopathological characteristics of the patients. For molecular studies, 5 mL of blood was collected and sent to the laboratory in a sterile Eppendorf tube.
| Variables | Good Prognosis | Poor Prognosis |
|---|---|---|
| Sample size | 50 | 50 |
| Gender | 21 females; 29 males | 21 females; 29 males |
| Age | 52.3 ± 4.7 | 51.8 ± 4.1 |
3.2. Preparation of Peripheral Blood Mononuclear Cell
One hundred SARS-CoV-2 patients were divided into two groups, and their whole blood and PBMCs (prepared using the Ficoll method) were collected. Peripheral blood samples (4 mL each) were drawn into vacutainer tubes. PBMCs were separated using a Ficoll density gradient centrifugation process. The blood was first diluted with 1 phosphate-buffered saline (PBS) at a 1:1 ratio and then transferred into Ficoll tubes. The buffy coat containing PBMCs was carefully pooled and transferred into a 15-mL Falcon tube after centrifugation (20 minutes, 1000x g, room temperature). The PBMCs were subsequently washed twice with 10 mL of PBS and centrifuged for 10 minutes at 250 g. Complete RNA was then extracted from the samples.
3.3. RNA Extraction
After the separation of WBCs, a specialized Exiqon microRNA extraction kit (Exiqon, Denmark) was employed to extract microRNA from the WBCs. The process began by homogenizing the sample in a designated buffer provided by the kit. Subsequently, RNA was extracted through the addition of alcohol and a purifying buffer, following the kit's protocol. This method ensured the effective isolation of high-quality microRNA for further analysis.
3.4. cDNA Syntheses and Real-time Quantitative Reverse Transcription-PCR
All mRNAs were converted to cDNAs using the stem-loop method with the TAKARA cDNA Synthesis Kit (TAKARA, Cat No. 6130) following the manufacturer’s protocol. To validate the extraction and conversion steps to cDNAs, the expression of the U6 gene, a type of housekeeping gene, was examined. Subsequently, the expression levels of the target miRNAs were quantified using real-time PCR. The results were statistically analyzed and compared between severe and mild COVID-19 cases. The stem-loop structures and primer sets used for the microRNAs are listed in Table 2.
| Primer | Sequence |
|---|---|
| U6 stem loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAATAT |
| U6 forward | GCTTCGGCAGCACATATACTAAAAT |
| U6 reverse | CGCTTCACGAATTTGCGTGTCAT |
| miR-26a stem loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAAGCCTA |
| miR-125b stem loop | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGATCACAA |
| miR-26a forward | GGGCCTTTCAAGTAATCC |
| miR-125b forward | ATCGCTTCCCTGAGACCC |
| Universal reverse | CCAGTGCAGGGTCCGAGGTA |
3.5. Statistical Analyses
GraphPad Prism (version 6.0 for Windows) was used for statistical analysis. The results supported the hypothesis that the two specimens originated from distinct populations. Data were expressed as the mean ± standard error or standard deviation. The normality of data distribution was evaluated using the Shapiro-Wilk and Kolmogorov-Smirnov tests. An independent t-test was employed to compare miR-26a and miR-125b expression levels between case and control samples. A P-value of less than 0.05 (P < 0.05) was considered statistically significant. Receiver operating characteristic (ROC) curve analysis was used to assess the diagnostic value of different variables between the two groups (as a binary classifier). The area under the ROC curve (AUC) indicated the predictive value of the variables between the two patient groups.
4. Results
4.1. Low Expression of the miR-26a and miR-125b in Patients with a Poor Prognosis
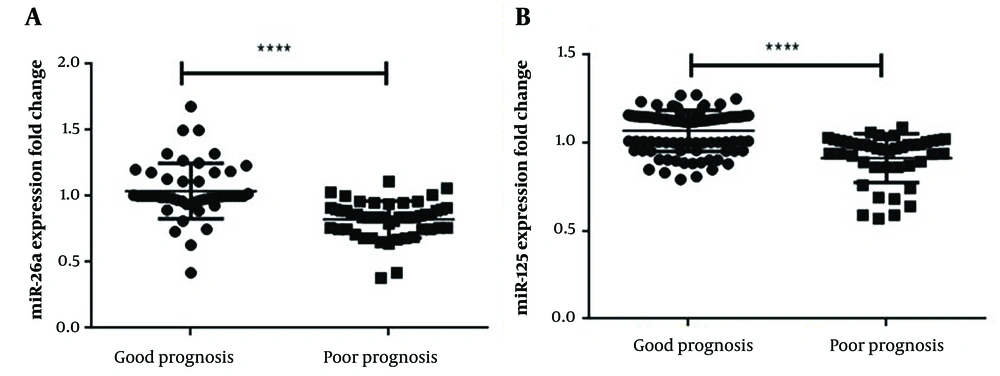
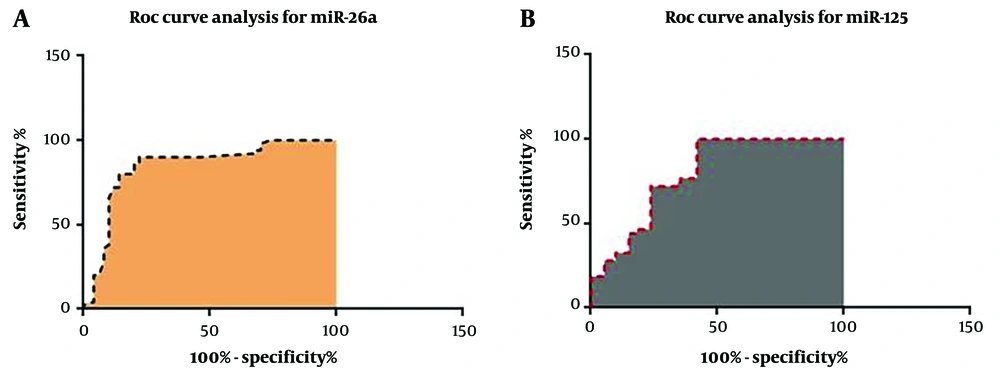
The qRT-PCR analysis data revealed that the PBMC levels of both miR-26a and miR-125b were significantly lower in patients with a poor prognosis compared to those with a good prognosis (P < 0.0001) (Figure 1). The clinical and pathological characteristics of the patients, including age and gender, were evaluated alongside the qRT-PCR results. Additionally, ROC curve analysis was employed to assess the specificity and sensitivity of miR-26a and miR-125b as potential indicators for the severity of SARS-CoV-2 infection. The biomarker index for miR-26a was 0.8458 (P < 0.0001), while the biomarker index for miR-125b was 0.7944 (P < 0.0001). The ROC analysis results are presented in Figure 2 and Table 3, while demographic information is detailed in Table 1.
Both the expression levels of miR-26a (A); and miR-125 (B) genes were significantly down-regulated in patients with a poor prognosis compared to patients with a good prognosis. Given the significant difference in miR-26a and miR-125 expression between patients with a good prognosis, it is proposed that future research may lead to the development of new therapeutic approaches for SARS-coV2 patients. The Student t-test was used to compare the statistical significance of the groups, and the results were normalized using the U6 housekeeping gene expression. **** P ≤ 0.0001.
| ROC Curve Data | miR-26a Values | miR-125 Values |
|---|---|---|
| Area | 0.8458 | 0.7944 |
| Standard error | 0.04235 | 0.03722 |
| 95% confidence interval | 0.7628 to 0.9288 | 0.7215 to 0.8674 |
| P-value | < 0.0001 | < 0.0001 |
| Controls (patient with good prognosis) | 50 | 43 |
| Patients (patient with poor prognosis) | 50 | 43 |
| Missing controls | 0 | 0 |
| Missing patients | 0 | 7 |
Abbreviation: ROC, receiver operating characteristic.
5. Discussion
SARS-CoV-2 infection can result in severe pulmonary disease, complications, and significant morbidity and mortality. Currently, there is no optimal therapy or effective medication for this potentially fatal lung condition. The host immune response to SARS-CoV-2 infection remains poorly understood, presenting challenges in the development of novel therapeutics. In many cases, viral infection triggers substantial changes in the host transcriptome, leading to abnormal host cell metabolism and a modulated immune response that supports viral replication (16). Understanding the molecular pathways involved in viral replication within human cells is critical for developing effective pharmacological strategies and identifying innovative biomarkers (17).
Biomarkers play a crucial role during the progression of this disease, particularly in classifying patients as mild, severe, or critical, thereby enabling earlier intervention for SARS-CoV-2 patients (18, 19). In patients with severe SARS-CoV-2 infection, a pattern of hematologic, metabolic, inflammatory, and immune-biological anomalies has been reported compared to those with milder systemic illnesses (17). Guterres et al. identified 34 miRNAs for positive-sense viral RNA and 45 miRNAs for negative-sense viral RNA that can strongly bind to key SARS-CoV-2 genes. These miRNAs hold significant potential in the immunopathogenesis and therapeutic approaches for SARS-CoV-2 disease (e.g., miR-18b, miR-193a, miR-367, and miR-668). According to their study, these miRNAs are also implicated in respiratory and cardiovascular diseases, such as lung cancer, asthma, pneumonia, and cardiac fibrosis (20).
We investigated miR-26a and miR-125b as potential biomarkers for classifying patients with mild versus severe symptoms. MiR-26a is involved in cancer and inflammation. Recent studies have demonstrated its role in asthma by regulating inflammatory factors and cells (11). MiR-26a also contributes to the GAS5-alleviated palmitic acid-induced myocardial inflammatory injury through the miR-26a/HMGB1/NF-κB axis (21). Moreover, miR-26a regulates pro-inflammatory cytokine production in microglia (9).
A study on influenza A virus (IAV) indicated that miR-26a expression significantly inhibits IAV replication by activating type I interferon (IFN) signaling pathways and promoting the expression of IFN-stimulated genes, thereby suppressing viral replication (22). Similarly, in the context of porcine reproductive and respiratory syndrome virus (PRRSV) infection, overexpression of the miR-26 family strongly inhibited PRRSV replication (23). Centa et al. observed that miR-26a, miR-29b, and miR-34a expression levels were significantly reduced in lung biopsies from COVID-19 patients compared to other lung diseases (24). Zhang et al. demonstrated that miR-26a promotes STAT1 phosphorylation during FHV-1 infection, which upregulates IFN-β expression. They also revealed that miR-26a stimulates IFN-I antiviral signaling, controls, and inhibits FHV-1 infection by targeting SOCS5, a negative regulator of the JAK-STAT signaling pathway (25).
NF-κB signaling regulates the expression of miR-125b, which inhibits the inflammatory response by targeting the TNF-α 3′UTR region gene (26). Busch et al. reported that miR-125b directly targets and inhibits the expression of a gene encoding 5-lipoxygenase, an essential enzyme in the biosynthesis of leukotrienes required for innate immune responses and inflammatory processes (15). In hepatitis B virus-infected cells, miR-125b expression was reduced, and ectopic expression of miR-125b inhibited HBV DNA intermediates, as well as HBsAg and HBeAg secretion. Additionally, miR-125b inhibited SCNN1A's mRNA and protein levels (27).
The angiotensin-converting enzyme 2 (ACE2) transmembrane protein on host cells plays a critical role in SARS-CoV-2 infection by acting as a receptor for the viral spike glycoprotein (28). Widiasta et al. revealed that miR-125b and miR-18 directly target the 3′UTR of ACE2 mRNA, suppressing ACE2 expression, which is crucial in the progression of COVID-19 infection (29). Our results show that the expression of both miR-26a and miR-125b is significantly decreased in patients with a poor prognosis compared to those with a good prognosis. These findings suggest that these miRNAs may serve as potential diagnostic markers for severe SARS-CoV-2 infections. Their association with specific clinicopathological features and roles in inflammation and immune response disorders, as evidenced by previous studies, further support their potential utility. However, additional research is necessary to confirm these findings.
One limitation of this study was the exclusive focus on miR-26a and miR-125b as biomarkers, without considering other microRNAs or clinical indicators, which could provide a more comprehensive diagnostic profile. Another limitation was the relatively small sample size (100 samples), which may affect the robustness and generalizability of the findings. Further studies with larger cohorts and an expanded range of biomarkers are needed to enhance the diagnostic and prognostic understanding of these miRNAs in SARS-CoV-2 infection.
5.1. Conclusions
Given the substantial global threat posed by SARS-CoV-2 as an ongoing pandemic, and the critical importance of biomarkers in guiding optimal treatment strategies, it is anticipated that future research into the interaction of SARS-CoV-2 with various human cells will pave the way for novel therapeutic, diagnostic, and prognostic approaches to prevent and manage this infectious disease. In summary, our findings suggest that the relative expression levels of miR-26a and miR-125b in SARS-CoV-2 infection can serve as valuable indicators of disease severity and as prognostic markers.