1. Background
Pseudomonas aeruginosa is a clinically important opportunistic pathogen with resistance to various antimicrobial agents (1). This microorganism is one of the leading causes of life-threatening infections, particularly in individuals with immune system deficiencies, such as those with cystic fibrosis, AIDS, cancer, burns, wounds, and in intensive care units (ICUs) (2). The emergence of multidrug-resistant isolates has become one of the most concerning global issues today (3). Carbapenems are bactericidal agents that act on the cell wall and are used against severe Pseudomonas infections, often as a last resort (4). These antimicrobials consist of a penem fused to a beta-lactam ring and inhibit peptidoglycan synthesis by binding to and inactivating penicillin-binding proteins (PBPs) (5). However, their effectiveness in infection management has decreased with the emergence of carbapenem-resistant P. aeruginosa (CRPA) strains (6).
Impaired permeability of the porin OprD due to loss or alteration, hyperexpression of an active efflux pump, or production of class B metallo-β-lactamases (MBLs) are the primary mechanisms responsible for the emergence of CRPA strains (7). Metallo-β-lactamases are zinc-dependent enzymes belonging to Ambler class B β-lactamases that hydrolyze all β-lactam antibiotics, especially carbapenems, with the exception of monobactams (aztreonam), and are inhibited by EDTA (ethylenediaminetetraacetic acid; a metal chelator) (8). There are three subclasses (B1, B2, and B3) of Ambler class B β-lactamases, with subclass B1 being the most common worldwide (9). The B1 MBL subclass, including the blaIMP, blaVIM, bLasIM, bLasPM, blaGIM, blaDIM, blaIND, and blaNDM families, has been identified in P. aeruginosa worldwide. Plasmid-carried MBL genes are particularly important due to their widespread global distribution (10).
A cell density-dependent cell-to-cell signaling mechanism known as quorum sensing (QS) is recognized in P. aeruginosa for controlling the expression of biofilm formation genes and virulence factors (11, 12). las and rhl are two major components of the QS system, encoding N-(3-oxododecanoyl)-L-homoserine lactone (3O-C12-HSL) and N-butanoyl-L-homoserine lactone (C4-HSL) as autoinducers, respectively (13). The rhl system contributes to various phases of biofilm production, including microcolony formation, maintenance of open-channel architecture, and detachment of embedded bacterial cells (14). Additionally, rhl controls P. aeruginosa virulence factors, such as rhamnolipids, pyocyanin, hydrogen cyanide, and elastase (12). The las system regulates other features of the biofilm production process, such as biofilm maturation, and also controls genes encoding elastase, exotoxin A, and alkaline protease (15).
Currently, various typing approaches, such as bio-typing, sero-typing, and pyocin-typing, have been established to determine common clones of P. aeruginosa (16). Identifying these clones and understanding their relationships is crucial in epidemiological studies of P. aeruginosa hospital infections, the global prevalence of these bacteria, and the determination of disease burden (17). However, the discriminatory power of these methods is lower compared to nucleic acid amplification-based typing techniques (18). Therefore, several molecular typing methods have been developed, including pulsed-field gel electrophoresis (PFGE), multiple-locus variable number tandem repeat (MLVA), ribotyping, enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR), and random amplification of polymorphic DNA (RAPD) (19).
Although PFGE has high sensitivity and is recognized as the gold standard genotyping technique, it is limited by technical complexity, cost, the need for trained personnel, and long turnaround times for results (20). One disadvantage of MLVA is that it is not a universal method, as primers must be designed for each specific application, and results cannot be directly compared with those from other laboratories due to differences in amplicon band patterns. One of the most widely used repetitive DNA targets in PCR-based genotyping approaches is the ERIC sequence, which is common to gram-negative enteric bacteria (21). ERIC sequences offer better potential for the bacterial interspersed repetitive sequence evolution project because they are longer, more informative in comparative analyses, and found in a wider range of species (22).
2. Objectives
This study was conducted to investigate the QS genes and molecular typing of CRPA strains using the ERIC sequence method.
3. Methods
3.1. Sample Collection and Bacterial Identification
In this cross-sectional study, conducted over one year (April 2022 to March 2023), a total of 57 non-duplicative CRPA isolates were collected from two large hospitals in Babol, northern Iran. P. aeruginosa strains were initially identified using microbiological and biochemical standard tests and then confirmed by species-specific primers (oprL) that amplify a 504 bp product (13). Carbapenem resistance was determined using the disk diffusion method with imipenem disks (IPM; 10 µg; susceptible ≥ 19 mm, intermediate 16 - 18 mm, resistant ≤ 15 mm; Padtan Teb Co., Iran). Pseudomonas aeruginosa ATCC 27853 was used as a control.
3.2. Carbapenemase Production Tests
The modified-carbapenem inactivation method (mCIM) and the EDTA-modified carbapenem inactivation method (eCIM) tests were performed to detect carbapenemase and metallo-β-lactamase activity in the isolates, respectively (23).
3.3. Microtiter Biofilm Formation Assay
A polystyrene flat-bottom microtiter plate was used for the biofilm formation assay. Briefly, 200 µL of fresh bacterial suspension in brain heart infusion broth was added to each well. After overnight incubation at 37°C, each well was washed twice with phosphate-buffered saline to remove detached, weakly attached, and floating 'planktonic' cells. The plates were shaken to eliminate all non-adherent bacteria and then desiccated at room temperature to fix the attached bacteria. Staining was performed with 200 µL of 0.1% crystal violet (Sigma, St. Louis, USA) for 5 minutes at 25°C, followed by gentle washing with water and drying. The optical density (OD 570 nm) was measured using a microplate ELISA reader (BioTek, Bad Friedrichshall, Germany). All tests were repeated three times, and the cut-off value (ODc) was determined. Biofilm formation ability was classified as follows: Non-biofilm forming (ODt < ODc), weakly forming (ODc < ODt < 2x ODc), moderately forming (2x ODc < ODt < 4x ODc), and strongly forming (ODt ≥ 4x ODc) (24).
3.4. Polymerase Chain Reaction
Template DNA was extracted from fresh colonies as follows: Four to five pure colonies were suspended in 25 µL of 0.25% sodium dodecyl sulfate (SDS)–0.05 N NaOH solution and heated for 12 minutes. Then, 200 µL of distilled water was added to each tube, and 5 µL of the diluted mixture was used in the PCR reaction. The genomic DNA quantity was evaluated using a Nanodrop spectrophotometer (ND-1000; Thermo Scientific; Wilmington, DE, USA). The primer sequences are shown in Table 1. Polymerase chain reaction was performed in a total volume of 12 µL, which included 1.0 µL of template DNA, 4.8 µL of pre-made MasterMix (Ampliqon, Stenhuggervej, Denmark), 5.3 µL of nuclease-free ddH2O, and 0.3 µL of each primer (at a concentration of 10 pmol/µL). The samples were amplified in a Techne TC-512 thermocycler (Eppendorf, Hamburg, Germany) with the following conditions: One cycle of initial amplification at 95°C for 9 minutes, followed by 32 cycles of denaturation (95°C for 30 seconds), annealing (58°C for 60 seconds), extension (72°C for 60 seconds), and concluding with a final extension at 72°C for 7 minutes. Polymerase chain reaction products were subjected to 1.5% agarose gel electrophoresis prepared in 1X TBE (Tris/Borate/EDTA) buffer, stained with SafeStain loading dye (CinnaGen Co., Iran), and photographed under ultraviolet illumination (Bio-Rad, Hercules, USA).
| Target Genes and Primer Sequences | Product Size (bp) | References |
|---|---|---|
| β-lactamase encoded genes | (25) | |
| IMP | 740 | |
| F: 5ʹ- TGAGCAAGTTATCTGTATTC-3ʹ | ||
| R: 5ʹ- TTAGTTGCTTGGTTTTGATG-3ʹ | ||
| VIM | 390 | |
| F: 5ʹ- GATGGTGTTTGGTCGCATA-3ʹ | ||
| R: 5ʹ- CGAATGCGCAGCACCAG -3ʹ | ||
| NDM | 475 | |
| F: 5ʹ- GGGCAGTCGCTTCCAACGGT-3ʹ | ||
| R: 5ʹ- GTAGTGCTCAGTGTCGGCAT-3ʹ | ||
| OXA-23-like | 501 | |
| F: 5ʹ- GATCGGATTGGAGAACCAGA -3ʹ | ||
| R: 5- ATTTCTGACCGCATTTCCAT -3ʹ | ||
| OXA-24/40-like | 246 | |
| F: 5ʹ- GGTTAGTTGGCCCCCTTAAA-3ʹ | ||
| R: 5ʹ - AGTTGAGCGAAAAGGGGATT-3ʹ | ||
| OXA-85-like | 599 | |
| F: 5ʹ- AAGTATTGGGGCTTGTGCTG-3ʹ | ||
| R: 5ʹ- CCCCTCTGCGCTCTACATAC-3ʹ | ||
| OXA-48-like | 744 | |
| F: 5ʹ- TTGGTGGCATCGATTATCGG-3ʹ | ||
| R: 5ʹ- GAGCACTTCTTTTGTGATGGC-3ʹ | ||
| KPC | 882 | |
| F: 5ʹ- ATGTCACTGTATCGCCGTCT-3ʹ | ||
| R: 5ʹ- TTACTGCCCGTTGACGCCC -3ʹ | ||
| QS genes | ||
| lasR | 725 | (11) |
| F: 5ʹ - CTGTGGATGCTCAAGGACTAC-3ʹ | ||
| R: 5 ʹ - AACTGGTCTTGCCGATGG -3ʹ | ||
| rhlI | 155 | (11) |
| F: 5- GTAGCGGGTTTGCGGATG-3ʹ | ||
| R: 5- CGGCATCAGGTCTTCATCG-3ʹ | ||
| rhlR | 133 | (11) |
| F: 5 ʹ - GCCAGCGTCTTGTTCGG-3ʹ | ||
| R: 5 ʹ - CGGTCTGCCTGAGCCATC-3ʹ | ||
| lasI | 605 | (11) |
| F: 5 ʹ - CGCACATCTGGGAACTCA-3ʹ | ||
| R: 5 ʹ - CGGCACGACGATCATCATCT-3ʹ | ||
| aprA | 140 | (11) |
| F: 5 ʹ -ACCCTGTCCTATTCGTTCC-3' | ||
| R: 5 ʹ -GATTGCAGCGACAACTTGG-3' | ||
| rhlAB | 151 | (26) |
| F: 5'- TCATGGAATTGTCACAACCGC-3' | ||
| R: 5'- ATACGGCAAAATCATGGCAAC-3' | ||
| aprR | 506 | (27) |
| F: 5 ʹ -GTGCTGACCCTGTCCTATTC-3ʹ | ||
| R: 5 ʹ -GTGTTCTGCTCTTCCCAGTAG-3ʹ |
Abbreviation: QS, quorum sensing.
3.5. Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction Method
A set of primers, ERIC1 (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC2 (5′-AAGTAAGTACTGGGGTGAGCG-3′), were used for ERIC typing (28). Polymerase chain reaction was performed in a total volume of 12 µL, which included 1.2 µL of genomic DNA, 5.0 µL of pre-made MasterMix (Ampliqon, Stenhuggervej, Denmark), 5.0 µL of nuclease-free ddH2O, and 0.4 µL of each primer (at a concentration of 10 pmol/µL). The ERIC genotyping thermal cycling conditions included an initial 5 minutes at 94°C, followed by 33 cycles consisting of denaturation at 94°C for 60 seconds, annealing at 53°C for 60 seconds, and extension at 72°C for 5 minutes, with a final extension at 72°C for 10 minutes. Amplicons were analyzed using electrophoresis through 1.5% agarose gels and visualized under ultraviolet light after staining with SafeStain loading dye (CinnaGen Co., Tehran, Iran). The GelJ software (V.2) was used to evaluate the ERIC profiles, as previously described. A similarity coefficient of 80% or higher was considered to indicate similar genotypes.
3.6. Statistical Analysis
The chi-square test was used to assess the relationship between biofilm formation, resistance genes, and QS genes. A P-value of < 0.05 was considered statistically significant.
4. Results
In this study, 57 non-repetitive CRPA isolates were collected from various clinical samples as follows: Sputum (22.8%, n = 13/57), BAL (21.1%, n = 12/57), wound (17.5%, n = 10/57), urine (14.0%, n = 8/57), blood (14.0%, n = 8/57), and stool (10.5%, n = 6/57). The frequency of CRPA strains according to hospital wards was ICU (36.8%, n = 21/57), Emergency (22.8%, n = 13/57), Internal (21.1%, n = 12/57), and Infectious (19.3%, n = 11/57). The carbapenemase test conducted with the mCIM assay showed that 89.5% (n = 51/57) of CRPA strains were positive for carbapenemase production. To differentiate MBLs from serine carbapenemase, both mCIM and eCIM phenotypic tests were used. Among the CRPA isolates, 17.5% (n = 10/57) were positive in the eCIM tests.
The biofilm formation test showed that out of the 57 CRPA isolates, 10.5% (n = 6/57), 19.3% (n = 11/57), and 70.2% (n = 40/57) had weak, moderate, and strong biofilms, respectively. The presence of β-lactamase-encoding genes revealed that 75.4% (n = 43/57), 64.9% (n = 37/57), 12.3% (n = 7/57), and 8.7% (n = 5/57) carried blaIMP, blaVIM, blaNDM, and blaKPC, respectively. On the other hand, 73.7% (n = 42/57), 7.0% (n = 4/57), and 1.7% (n = 1/57) of CRPA isolates carried blaOXA-48-like, blaOXA-23-like, and blaOXA-20/40-like, respectively. No isolates were found to carry blaOXA-58-like. Table 2 shows the correlation between β-lactamase-encoded genes and biofilm production. No significant correlation was observed between the β-lactamase-encoded genes and biofilm formation ability, except for the blaKPC gene.
The molecular distribution of QS genes showed that 100% (n = 57/57), 96.5% (n = 55/57), 92.9% (n = 53/57), 89.5% (n = 51/57), 84.2% (n = 48/57), 73.6% (n = 42/57), and 63.2% (n = 36/57) of isolates harbored the lasR, lasI, rhlI, rhlR, aprR, aprA, and RhlAB genes, respectively (Table 3). Table 4 shows that isolates harboring QS genes produced stronger biofilms than those without these genes. However, no significant correlation was observed between the QS genes and biofilm formation ability.
| Antibiotic Resistance Genes | No. of Isolates | Biofilm Strength | P-Value | ||
|---|---|---|---|---|---|
| Weak (N = 6) | Moderate (N = 11) | Strong (N = 40) | |||
| blaIMP | 43 (75.4) | 4 (8.9) | 9 (20) | 32 (71.1) | 0.731 |
| blaVIM | 37 (64.9) | 3 (8.1) | 6 (16.2) | 28 (75.7) | 0.459 |
| blaNDM | 7 (12.3) | 3 (42.9) | 0 | 4 (57.1) | 0.008 |
| blaKPC | 5 (8.7) | 1 (20) | 1 (20) | 3 (60) | 0.76 |
| blaOXA-48-like | 42 (73.7) | 5 (11.9) | 7 (16.7) | 30 (71.4) | 0.639 |
| blaOXA-23-like | 4 (7) | 0 | 1 (25) | 3 (75) | 0.764 |
| blaOXA-20/40-like | 1 (1.7) | 0 | 0 | 1 (100) | 0.805 |
a Values are expressed as No. (%).
| Strain Number | Source of Samples | Clinical Wards | mCIM | eCIM | BF | Resistance Genes | QS-Encoded Genes |
|---|---|---|---|---|---|---|---|
| 1 | Urine | Emergency | + | - | Strong | IMP/ VIM/ NDM | lasR/ lasI/ rhlR/ aprR / aprA/ rhlAB |
| 2 | Wound | Infectious | + | + | weak | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR/ rhlAB |
| 3 | Stool | Internal | + | - | Strong | VIM/ NDM/ OXA-48-like | lasR/ lasI/ rhlI/ aprR / aprA |
| 4 | BAL | ICU | + | - | Moderate | IMP/ KPC/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR/ rhlAB |
| 5 | BAL | Infectious | - | - | Strong | VIM/ NDM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR / aprA |
| 6 | Blood | ICU | + | - | Strong | IMP/ VIM | lasR/ lasI/ rhlI/ aprR/ aprA/ rhlAB |
| 7 | BAL | Infectious | + | + | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprA |
| 8 | Blood | ICU | + | - | Weak | NDM/ KPC/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR/ rhlAB |
| 9 | Urine | Internal | - | - | Moderate | VIM/ OXA-23-like | lasR/ lasI/ rhlI/ rhlR/aprR / aprA |
| 10 | Sputum | ICU | + | - | Strong | IMP/ VIM | lasR/ lasI/ rhlI/ rhlR/aprR/ rhlAB |
| 11 | Urine | Infectious | + | - | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprA |
| 12 | Wound | Internal | + | - | Strong | IMP/ NDM/ OXA-48-like | lasR/ lasI/ rhlR/ aprR/ aprA/ rhlAB |
| 13 | BAL | ICU | + | - | Moderate | VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR/ AprA/RhlAB |
| 14 | Urine | Emergency | + | - | Weak | IMP/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR / aprA/ rhlAB |
| 15 | BAL | ICU | + | - | Strong | IMP/ VIM | lasR/ lasI/ rhlI/ aprR/ aprA |
| 16 | Stool | Internal | + | - | Weak | VIM/ NDM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR/ rhlAB |
| 17 | Blood | ICU | - | + | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprA/ rhlAB |
| 18 | Stool | Infectious | + | - | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR/ rhlAB |
| 19 | Urine | Emergency | + | - | Strong | IMP/ VIM | lasR/ lasI/ rhlI/ rhlR/ aprA/ rhlAB |
| 20 | Sputum | ICU | + | - | Strong | IMP/ VIM | lasR/ rhlI/ rhlR/ aprR / aprA/ rhlAB |
| 21 | Stool | Emergency | + | - | Strong | VIM/ OXA-48-like | lasR/ lasI /rhlIrhlR/aprR/ aprA |
| 22 | BAL | ICU | - | + | Moderate | IMP/ VIM | lasR/ lasI /rhlI/ rhlR/aprR/ aprA |
| 23 | Sputum | Internal | + | - | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlR/ aprR / aprA/ rhlAB |
| 24 | Urine | Emergency | + | - | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlR/ aprR / aprA |
| 25 | Stool | ICU | + | - | Moderate | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlR/ aprR / aprA |
| 26 | Sputum | Infectious | + | - | Strong | KPC/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprA/ rhlAB |
| 27 | Sputum | ICU | + | + | Strong | IMP/ OXA-23-like/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR / rhlAB |
| 28 | BAL | ICU | + | - | Weak | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ aprR/ aprA |
| 29 | Urine | Infectious | + | - | Moderate | IMP/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR/ aprA |
| 30 | Stool | Infectious | + | - | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR/ aprA/ rhlAB |
| 31 | BAL | ICU | + | - | Strong | VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR/ rhlAB |
| 32 | Sputum | Internal | - | - | Moderate | IMP/ VIM | lasR/ lasI/ rhlI/ rhlR/aprR / aprA |
| 33 | Sputum | ICU | + | - | Moderate | IMP/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR / rhlAB |
| 34 | Sputum | ICU | + | - | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprA/ rhlAB |
| 35 | Urine | Emergency | + | + | Moderate | IMP/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR / aprA/ rhlAB |
| 36 | BAL | Infectious | + | - | Strong | VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR / aprA/ rhlAB |
| 37 | Sputum | Internal | + | - | Strong | IMP/ OXA-48-like | LasR/ LasI/ RhlI/ RhlR/ aprR / AprA |
| 38 | Sputum | Internal | + | - | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ aprR / aprA/ rhlAB |
| 39 | Sputum | Internal | + | - | Strong | KPC/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR/ aprA |
| 40 | BAL | ICU | + | - | Strong | IMP/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR/ rhlAB |
| 41 | Wound | Emergency | + | - | Strong | IMP/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR / aprA |
| 42 | Sputum | Internal | + | - | Strong | IMP/ VIM | lasR/ lasI/ rhlI/ rhlR/ aprA/ rhlAB |
| 43 | Wound | Internal | + | - | Strong | IMP/ VIM | lasR/ lasI/ rhlI/ rhlR/aprR / aprA/ rhlAB |
| 44 | Sputum | Infectious | + | + | Strong | IMP/VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/aprR / rhlAB |
| 45 | BAL | ICU | + | - | Moderate | IMP/VIM | lasR/ lasI/ rhlI/ rhlR/aprR/ aprA |
| 46 | Wound | Emergency | + | - | Strong | IMP/ VIM | lasR/ lasI/ rhlI/ rhlR/ rhlAB |
| 47 | Wound | Infectious | + | - | Strong | IMP/ OXA-48-like | lasR/ lasI / rhlI/ rhlR/aprR/ aprA/ rhlAB |
| 48 | Wound | Emergency | + | - | Weak | IMP/ NDM | lasR/ lasI/ rhlI/ rhlR/ aprR/ aprA/ rhlAB |
| 49 | Blood | ICU | + | - | Strong | VIM/ KPC/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR/ aprA/ |
| 50 | BAL | ICU | + | - | Strong | IMP/ VIM/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR/ aprA/ rhlAB |
| 51 | Wound | Emergency | - | - | Moderate | IMP/ OXA-23-like/ OXA-48-like | lasR/ rhlI/ rhlR/ aprR/ aprA/ rhlAB |
| 52 | Blood | Emergency | + | + | Strong | IMP/ OXA-48-like | lasR/ lasI /rhlI/ rhlR/aprR |
| 53 | Blood | ICU | + | + | Strong | IMP/ OXA-48-like | lasR/ lasIrhlI/ rhlR/aprR / aprA/ rhlAB |
| 54 | Wound | Emergency | + | - | Strong | VIM/ OXA-24/40-like | lasR/ lasI/ rhlR/ aprR/ aprA |
| 55 | Blood | Emergency | + | - | Strong | IMP/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprA/ rhlAB |
| 56 | Blood | Internal | + | - | Strong | IMP/ OXA-48-like | lasR/ lasI/ rhlI/ rhlR/ aprR |
| 57 | Wound | ICU | + | + | Strong | VIM/ OXA-23-like/ OXA-48-like | lasR/ lasI/ rhlI/ aprR/ aprA/ rhlAB |
Abbreviations: BAL, Bronchoalveolar lavage; BF, biofilm formation; ICU, Intensive Care Unit; mCIM, modified-carbapenem inactivation method; eCIM, EDTA-modified carbapenem inactivation method; QS, Quorum sensing; CRPA, Carbapenem-resistant Pseudomonas aeruginosa.
| QS Genes | No. of Isolates | Biofilm Strength | P-Value | ||
|---|---|---|---|---|---|
| Weak (N = 6) | Moderate (N = 11) | Strong (N = 40) | |||
| lasR | 57 (100) | 6 (10.5) | 11 (19.3) | 40 (70.2) | - |
| lasI | 55 (96.5) | 3 (8.1) | 6 (16.2) | 28 (75.7) | 0.805 |
| rhlI | 53 (92.9) | 6 (11.3) | 11 (20.8) | 36 (67.9) | 0.401 |
| rhlR | 51 (89.5) | 5 (9.8) | 11 (21.6) | 35 (68.6) | 0.427 |
| aprR | 48 (84.2) | 6 (12.5) | 10 (20.8) | 32 (66.7) | 0.362 |
| aprA | 42 (73.6) | 3 (7.1) | 9 (21.4) | 30 (71.4) | 0.342 |
| rhlAB | 36 (63.2) | 5 (13.9) | 5 (13.9) | 26 (70.2) | 0.274 |
Abbreviation: QS, quorum sensing.
a Values are expressed as No. (%).
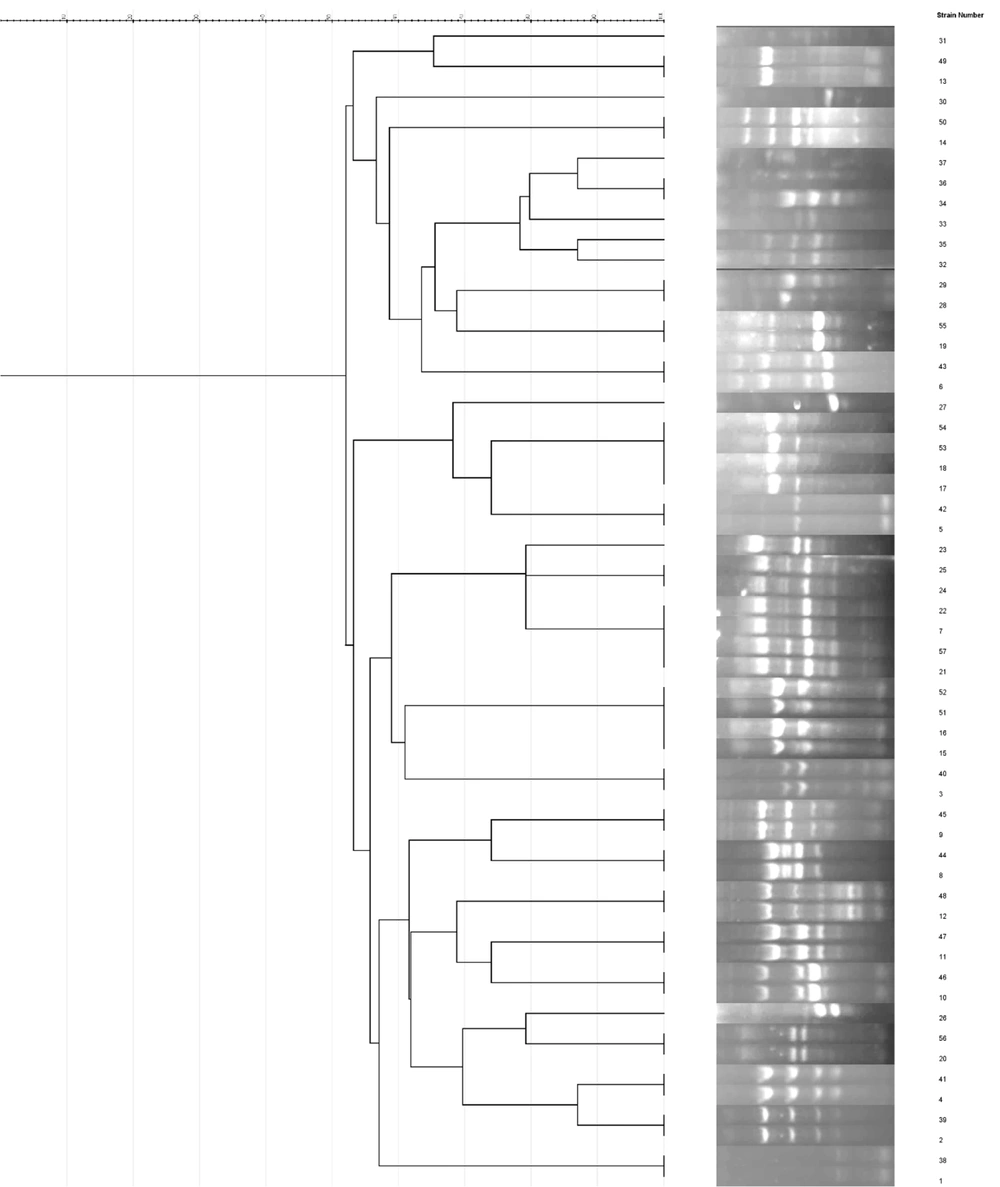
Enterobacterial repetitive intergenic ERIC-PCR analysis of 57 CRPA isolates identified eight distinct clusters (A, B, C, D, E, F, G, and H) using a similarity cut-off of ≥ 60%. The distribution of isolates within these clusters was as follows: A, (3 isolates); B, (2 isolates); C, (12 isolates); D, (7 isolates); E, (7 isolates); F, (6 isolates); G, (17 isolates); and H, (2 isolates). Additionally, one isolate was classified as a singleton. Notably, the G cluster exhibited the highest isolate count, with 17 strains (Figure 1).
5. Discussion
It is well established that P. aeruginosa infections are associated with high mortality and morbidity due to its ability to adapt to various environmental conditions, express many virulence factors, and acquire antimicrobial resistance (AMR) (11). In the present study, of the total 57 CRPA strains, 70.2%, 19.3%, and 10.5% were strong, moderate, and weak biofilm formers, respectively. Additionally, 89.5% and 17.5% of the isolates were positive in the mCIM and eCIM tests, respectively. The molecular distribution of β-lactamase-encoded genes revealed that 75.4%, 64.9%, 12.3%, and 8.7% of isolates carried blaIMP, blaVIM, blaNDM, and blaKPC, respectively. Furthermore, 73.7%, 7.0%, and 1.7% of isolates harbored blaOXA-48-like, blaOXA-23-like, and blaOXA-20/40-like, respectively. The blaOXA-58-like gene was not found in any of the isolates. In contrast to our findings, Vaez et al. showed that the prevalence of blaVIM, blaIMP, and blaNDM was 20% (18 of 90), 8.9% (8 of 90), and 5.6% (5 of 90) of P. aeruginosa isolates, respectively. They stated that the MBL genes (blaVIMand blaIMP) seem to play a critical role in the spread of carbapenem-resistant infections and the emergence of multidrug-resistant strains, leading to therapeutic failure.
In a study conducted by Ramazani et al., out of 117 non-duplicative P. aeruginosa isolates, the distribution of MBL genes was as follows: blaIMP (62.1%), blaVIM (31.0%), and blaNDM (6.8%) (25). This difference may be due to the origin of the samples, as most of the strains in the present study were multidrug-resistant (data not shown) and strong biofilm producers. Esmaeili et al. found that of 150 clinical samples, 75 P. aeruginosa isolates were recovered, and 68.6% were MBL producers, all of which carried the VIM gene (100%) (29). In a study by Doosti et al., 70 P. aeruginosa isolates were analyzed, and 56% (n = 23/41) and 24.3% (n = 10/41) carried the blaVIM and blaIMP genes, respectively (30). Dogonchi et al. reported that 90%, 40%, and 5% of CRPA isolates harbored the blaIMP, blaVIM, and blaNDM genes, respectively (31).
In line with our data, Ramazani et al. found that 75.8% (n = 22/29), 10.3% (n = 3/29), and 3.4% (n = 1/29) of CRPA isolates harbored blaOXA-48-like, blaOXA-23-like, and blaOXA-20/40-like, respectively (25). The blaOXA-58-like gene was not found in any isolate. In our study, and in agreement with Devanga Ragupathi et al., the distribution of resistance-coding genes in strains with strong biofilm formation was significantly different compared to other strains (32). Devanga Ragupathi et al. showed that 27.8% (n = 20/72) were strong biofilm producers, with a positive correlation to carbapenem resistance, compared to 22.2% moderate, 19.4% weak, and 30.6% non-biofilm formers (32). In agreement with our results, Rahdar et al. highlighted that the increased carbapenem resistance burden in biofilm-producing strains is a significant concern, and the implementation of basic measures to address this phenomenon is imperative (33).
The distribution of QS genes showed that 100%, 96.5%, 92.9%, 89.5%, 84.2%, 73.6%, and 63.2% of isolates carried the lasR, lasI, rhlI, rhlR, aprR, aprA, and rhlAB genes, respectively. In contrast, Salehi et al. reported that the frequencies of the rhlR, lasR, and lasI genes were 5%, 48.3%, and 60%, respectively, while the rhlI, lasB, apr, and rhlAB genes could not be identified in any of the strains (26). Ghanem et al. found that, out of 125 clinical isolates of P. aeruginosa, the frequencies of the lasB and aprA genes were 89.6% and 85.6%, respectively (11). Baskan et al. showed that two QS system genes were detected in 98.1% (n = 51/52), and co-existence of the four QS system genes (lasI/R and rhlI/R genes) was found in 78.8% (n = 41/52) of the isolates (34).
Interestingly, and in line with Karami et al., the presence of QS genes was significantly higher in strains with strong biofilm formation compared to other strains (P < 0.05) (35). Karami et al. also observed a significant correlation between the rhlA gene and biofilm formation capability (P = 0.02) (35). El Askary showed that, of 62 P. aeruginosa isolates, 70.9% were biofilm producers (36). The QS genes rhlI and lasI were present in 67.7% and 48.4%, respectively. The prevalence of the blaIMP and blaVIMgenes was found in 33.9% and 24.2% of CRPA isolates, respectively. In line with our data, there was a high propensity for biofilm production in P. aeruginosa, which increased with the presence of QS genes. Contrary to our study, no significant relationship was observed between biofilm formation and carbapenem resistance. This difference may result from geographical diversity, strain genetics, and differences in methodology.
Enterobacterial repetitive intergenic consensus typing revealed eight distinct clusters. The distribution of isolates within these clusters was as follows: A, (3 isolates); B, (2 isolates); C, (12 isolates); D, (7 isolates); E, (7 isolates); F, (6 isolates); G, (17 isolates); and H, (2 isolates). Additionally, one isolate was classified as a singleton. In line with our data, Eladawy et al. demonstrated that ERIC typing is an effective tool for detecting the similarity percentage between P. aeruginosa isolates (37). Hematzadeh and Haghkhah reported that, according to the Dice similarity coefficient of > 80%, 38 ERIC types were identified (16). Zarei et al. found 14 different ERIC fingerprints, including five unique types and nine common types, among P. aeruginosa isolates collected from clinical, environmental, and cockroach samples (38). In agreement with our study, Zarei et al. stated that ERIC-PCR is inexpensive, easy to perform, and provides reliable, rapid, and discriminatory power for typing P. aeruginosa (38).
5.1. Conclusions
This study demonstrates that CRPA isolates exhibit a high prevalence of strong biofilm formation and QS genes, which contribute significantly to their virulence and antibiotic resistance. The genetic diversity observed among CRPA strains, particularly in biofilm-associated and resistance genes, underscores the adaptability of these pathogens in clinical settings. These findings emphasize the need for targeted strategies to disrupt biofilm formation and QS pathways as potential approaches for controlling CRPA infections and mitigating treatment challenges in healthcare environments.