1. Background
The COVID-19 pandemic continues to be a global health concern. The genome of SARS-CoV-2 undergoes dynamic evolution. The WHO has identified five variants of concern (VOCs) (1). The presence of VOCs may pose challenges to current detection systems (2-5). The accuracy of nucleic acid testing is crucial for effective epidemic control (6, 7). The SARS-CoV-2 nucleic acid detection kits primarily target specific genes, including S, E, N, and RdRp (6). The cycle threshold (Ct) values obtained from detection kits using PCR instruments are related to the initial viral RNA load of SARS-CoV-2 in the sample (8, 9). In the early days of the COVID-19 pandemic, various SARS-CoV-2 nucleic acid detection systems were introduced to the market to meet the demand for clinical screening. The efficacy and clinical application of different detection systems vary depending on the principles of detection and technical approaches.
Many laboratories prefer RNA extraction-free SARS-CoV-2 nucleic acid assay kits due to their specific advantages and the convenience they offer in various scenarios. These assays provide benefits such as rapid turnaround time and simple workflow, making them suitable for certain situations (10, 11). Some studies have demonstrated the feasibility of extraction-free SARS-CoV-2 testing, showing largely concordant results between extraction-free RT-PCR testing and RT-PCR using extracted RNA. However, other studies have indicated that extraction-free protocols exhibit lower analytical sensitivity compared to conventional protocols. Consequently, most false-negative results are observed in samples with low viral loads. Additionally, different assays show variations in Ct values and inconsistent qualitative results (12-15).
2. Objectives
The present study aimed to evaluate the applicability of the kits in clinical settings and provide insights into future application scenarios, result analysis, and reagent optimization.
3. Methods
3.1. Standard and Clinical Samples
Low-concentration quality control material (Z0) and phage pseudovirus particles of SARS-CoV-2 were obtained from BDS Biological Company, China. These particles contained the full length of the RdRp, E, and N genes of SARS-CoV-2, along with partial sequences of the ORF1ab gene. The average concentration was 1.03 × 10³ copies/mL. The SARS-CoV-2 standard reference material (S0) (BDS Biological Company, China) consisted of SARS-CoV-2 pseudovirus particles, encompassing the full-length sequence of SARS-CoV-2, with an average concentration of 2.0 × 10⁵ copies/mL.
A respiratory multiplex nucleic acid assay quality control product (BDS Biological Company, China) was developed using cultures of influenza A virus (IVA), influenza B virus (IVB), respiratory syncytial virus (RSV), adenovirus (ADV), rhinovirus (RhV), and Mycoplasma pneumoniae (MP). The concentrations of these pathogens ranged from 1 × 103 copies/mL to 1 × 104 copies/mL. The SARS-CoV-2 variant samples (G1 - G4) were obtained from the National Center for Clinical Laboratory, China. The G1 represented the Omicron BA.5 variant (3.0 × 103 copies/mL), G2 contained the Omicron BF.7 variant (7.5 × 102 copies/mL), G3 included the Omicron BA.5 variant (7.5 × 102 copies/mL), and G4 contained the Omicron BF.7 variant (3.0 × 103 copies/mL). All positive and negative specimens were obtained from Wuxi People’s Hospital through nasopharyngeal swabs.
3.2. SARS-CoV-2 RT-PCR Nucleic Acid Detection Kits
Four different kits are available for the detection of SARS-CoV-2 through RT-PCR. Three of these kits, namely SX002 (Sansure Biotech, China), SX08 (Sansure Biotech, China), and KYD (Coyote Biotech, China), utilize RNA extraction-free methods. The SS kit (Bioperfectus Technology, China) employs a magnetic bead-based method for RNA extraction. Basic information about these kits is provided in Appendix 3 in Supplementary File.
3.3. Laboratory Procedure and Supporting Instruments of the Four RT-PCR Kits
3.3.1. SX002-QuantStudioTM Dx Assay
A total of 10 μL of the sample and 10 μL of the sample release agent were directly added to a PCR reaction tube. The reaction system was then amplified using the QuantStudioTM Dx PCR instrument (Thermo Fisher Scientific, USA).
3.3.2. SX08-MA-1630Q
The sample pre-treatment procedure is similar to that of the SX002 reaction system. The reaction system was amplified using the MA-1630Q PCR instrument (Xunrui Biotechnology, China).
3.3.3. KYD-Flash20
A total of 15 μL of the sample and 15 μL of the sample processing solution were added to a centrifuge tube. From this mixture, 15 μL was transferred to a special butterfly-shaped PCR reaction tube containing 37 μL of the PCR reaction mixture. PCR amplification was performed using the Flash20 instrument (Coyote Biotechnology, China).
3.3.4. SS-QuantStudioTM Dx
RNA was extracted from 200 μL of the sample using the GeneRotex96 automatic nucleic acid extractor (Tianlong Biotechnology, China). The PCR amplification was performed using the QuantStudioTM Dx PCR instrument.
3.4. Comparison of Linear Quantification
The S0 was diluted using a two-fold gradient, resulting in a series of concentration gradient plates: S1 (1:2) to S9 (1:512). These dilutions were tested twice using the four SARS-CoV-2 nucleic acid detection kits. The logarithm of the theoretical dilution concentration was plotted on the horizontal axis, while the mean Ct value of the test results was plotted on the vertical axis. A linear regression equation was obtained by drawing a fitting line, and the slope (K) and determination coefficient (R²) were calculated. The correlation among the Ct values for the target genes detected by different kits was compared.
3.5. Comparison of Precision
The S0 was diluted in a 10-fold gradient to Y1 (2.0 × 104 copies/mL). For intra-batch precision, analyses of the two concentrations (Y1 and Z0) were repeated 20 times within a single batch on the same day. For inter-batch precision, the two concentrations were tested five times per day for four consecutive days, totaling 20 tests. The Ct values for the target genes were used to calculate the mean values, standard deviation (SD), and coefficient of variation (CV) as measures of precision.
3.6. Comparison of Accuracy
The linear regression equations derived from the fitting curves of the four kits were used as quantitative standard curves for calculating the target gene concentration in the in-batch test results of samples Y1 and Z0. Corresponding concentrations were determined by inputting the Ct values obtained from the tests into these equations.
3.7. Comparison of Limit of Detection
The Z0 was subjected to serial dilution to obtain four dilution levels: Z1 (1:2) to Z4 (1:16). The corresponding theoretical concentrations were 500, 250, 125, and 62.5 copies/mL. Samples at each concentration were tested four times. The lowest concentration that consistently yielded a 100% positive detection rate across the four replicates was considered the limit of detection (LOD).
3.8. Analysis of Cross-reaction
Respiratory disease-related pathogens were tested to evaluate the specificity of each kit and to identify any potential cross-reactions.
3.9. Analysis of Detection Results
Thirty-five positive and twenty negative samples were tested using different kits. The SARS-CoV-2 quality control materials containing different Omicron variants were also detected using the four kits. The coincidence rate of the results was compared and analyzed. Additionally, the concentration and purity of nucleic acids in the samples, which were extracted using magnetic beads, were assessed.
4. Results
4.1. Comparison of Linear Quantification
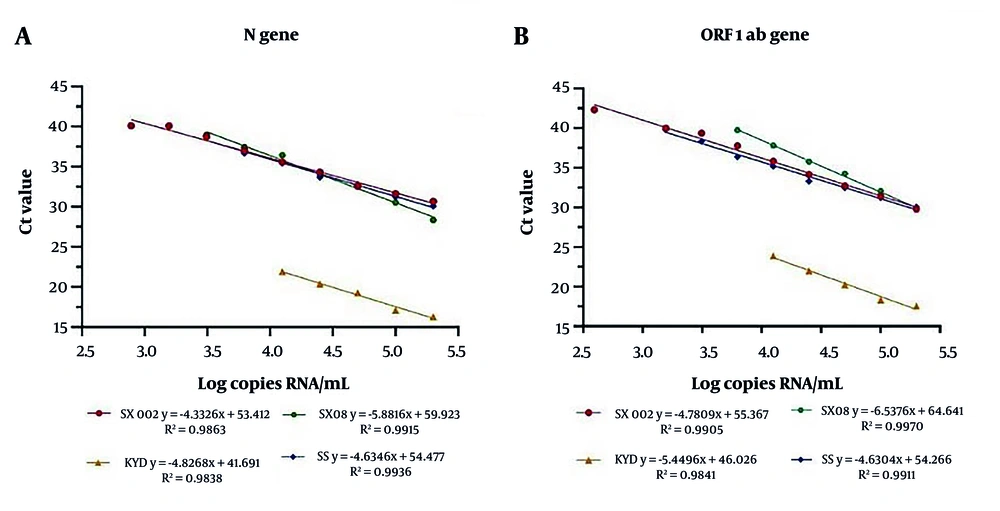
The logarithms of the theoretical concentrations of S0 - S9 were set as the X-axis, and the Ct values for the target genes detected were set as the Y-axis. The fitting regression lines are shown in Figure 1. The corresponding R² values for the SX002, SX08, KYD, and SS kits were 0.9863, 0.9915, 0.9838, and 0.9936 for the N gene, and 0.9905, 0.9970, 0.9841, and 0.991 for the ORF1ab gene, respectively. The correlation among the Ct values for the two target genes is illustrated in Appendix 1 in Supplementary File. The r-values for SX002, SX08, KYD, and SS were 0.994, 0.997, 0.995, and 0.999, respectively.
4.2. Comparison of Precision
Appendix 4 in Supplementary File presents the results of the intra-batch and inter-batch precision analyses for Y1 and Z0. The CV values were less than 5% for all intra-batch and inter-batch measurements. The SS kit exhibited the lowest intra-batch and inter-batch CV values (< 1%) for both target genes, indicating superior precision. In contrast, the KYD kit displayed the highest CV values for both target genes, reflecting greater variability.
4.3. Comparison of Accuracy
Appendix 5 in Supplementary File presents the accuracy assessment results for Y1 and Z0. The deviation was considered acceptable if the absolute deviation of the reference test results did not exceed ± 0.5 log orders of magnitude. For the Y1 concentration, the results of all four kits met the deviation requirements. The absolute deviation of the SS kit was less than 0.1 log, indicating high accuracy. The SX002 exhibited the smallest deviation of 0.037 for the N gene, while SX08 had the smallest deviation of 0.089 for the ORF1ab gene. The deviation of the test results was larger for the Z0 concentration, indicating reduced accuracy at lower concentrations.
4.4. Comparison of Limit of Detection
Table 1 displays the results of SARS-CoV-2 detection at various concentrations. The positive detection rates of SX002, SX08, and SS were 100% for both target genes at concentrations Z1 and Z2. Additionally, the N gene of KYD showed a 100% detection rate at these concentrations. At concentration Z3, SX002 had detection rates of 50% for the N gene and 75% for the ORF1ab gene, while SX08 showed a 75% detection rate for both target genes. The SS kit exhibited a 75% detection rate for both target genes, and KYD had a 75% detection rate for the N gene. At concentration Z4, SX002 failed to detect both target genes, whereas SX08 failed to detect the N gene only. The ORF1ab gene was detected at a rate of 25% for SX002 and 100% for SS. The N gene detection rate was 25% for both SX08 and KYD.
| Assay kits and Target Genes | Z1 | Z2 | Z3 | Z4 |
|---|---|---|---|---|
| SX002 | ||||
| N | √√√√ | √√√√ | √√ | Nd |
| ORF1ab | √√√√ | √√√√ | √√√ | Nd |
| SX08 | ||||
| N | √√√√ | √√√√ | √√ | Nd |
| ORF1ab | √√√√ | √√√√ | √√√ | √ |
| SS | ||||
| N | √√√√ | √√√√ | √√√ | √ |
| ORF1ab | √√√√ | √√√√ | √√√ | √√√√ |
| KYD | ||||
| N | √√√√ | √√√√ | √√√ | √ |
| ORF1ab | Nd | Nd | Nd | Nd |
The Lower Limit of Detection of Four Assay Kits
4.5. Analysis of Cross-Reaction
None of the kits showed specific amplification of non-target genes, indicating that all assays had good detection specificity.
4.6. Detection of SARS-CoV-2 Variant
The results are presented in Table 2 and Appendix 2 in Supplementary File. The SX002, SX08, and SS assays successfully detected both variants across the tested concentrations. However, the KYD kit did not exhibit specific amplification for the BA.5 variant at a concentration of 7.5 × 102 copies/mL.
| Assay Kits | N Gene | ORF1ab Gene | ||||||
|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G1 | G2 | G3 | G4 | |
| SX002 | 35.93 | 36.98 | 36.01 | 34.67 | 33.38 | 35.4 | 34.22 | 33.22 |
| SXO8 | 37.44 | 36.42 | 36.01 | 34.3 | 31.92 | 34.59 | 33.44 | 31.97 |
| SS | 37.41 | 40 | 38.41 | 36.58 | 36.86 | 39.35 | 37.46 | 37.79 |
| KYD | 23.17 | 22.73 | 22.02 | 19.39 | 21.63 | 23.38 | Nd | 24.14 |
The Results (Ct Values) of the Four Kits for the Omicron Variants
4.7. Analysis of Detection Results
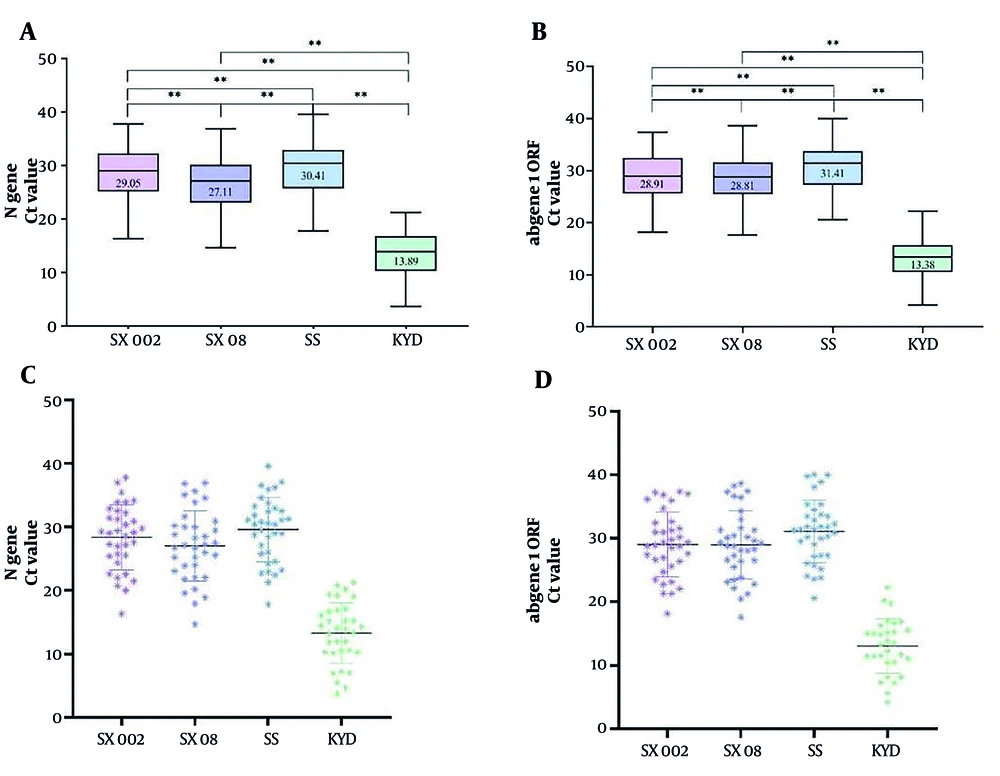
Thirty-five positive samples were detected. The Ct values (mean ± SD) for the N gene using SX002, SX08, KYD, and SS were 28.36 ± 5.13, 27.02 ± 5.53, 13.27 ± 4.76, and 29.57 ± 5.06, respectively. For the ORF1ab gene, the Ct values for SX002, SX08, KYD, and SS were 29.03 ± 5.09, 28.97 ± 5.36, 13.04 ± 4.31, and 31.08 ± 4.94, respectively. The SX08 and SS reagents tested positive in all positive specimens, while SX002 tested positive in 34 specimens, and KYD tested positive in 30 specimens. The detailed results are provided in Appendix 6. The Ct values obtained from KYD were consistently lower than those from the other reagents (P < 0.01, Figure 2).
The Ct values comparison of the four assay kits for the detection of positive samples. A and B, show the maximum, minimum, median, and quartile Ct values for each assay kit. The significance of the differences is denoted by **P < 0.01; C and D, depict scatter plots, illustrating the results obtained from the detection of the two target genes using the four reagents.
For the negative specimens, all four kits produced negative results for the target genes. The comparison of the three extraction-free detection kits with the SS kit showed that SX08 demonstrated 100% compliance with a κ value of 1, indicating perfect agreement. The KYD kit showed 90.91% compliance with a κ value of 0.814, indicating substantial agreement (Table 3). The purity values (mean ± SD) of the negative and positive samples were 1.91 ± 0.12 and 1.94 ± 0.14, respectively. The RNA with a purity range of 1.8 - 2.0 demonstrated high quality, and the purity was not significantly different between the negative and positive groups.
| SS | SX002 | SX08 | KYD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | OA | Positive | Negative | Total | OA | Positive | Negative | Total | OA | |
| Positive | 34 | 1 | 35 | 98.18% | 35 | 0 | 35 | 100% | 30 | 5 | 35 | 90.91% |
| Negative | 0 | 20 | 20 | κ = 0.961 (1.035 - 0.887) | 0 | 20 | 20 | κ = 1.000 (1.000 - 1.000) | 0 | 20 | 20 | κ = 0.814 (0.967 - 0.661) |
| Total | 34 | 21 | 55 | 35 | 20 | 55 | 30 | 25 | 55 | |||
Overall Agreement Between the Different Methods of Assessing SARS-CoV-2 RNA a
5. Discussion
Several studies have reported comparative analyses of the detection performance indicators of different SARS-CoV-2 nucleic acid kits. Results have shown that the efficiency of various nucleic acid extraction methods can affect test outcomes, and the Ct values obtained from different detection kits can vary significantly. In addition, some kits may fail to detect samples with low viral loads (6, 16-18). This study investigated the linear quantification of four SARS-CoV-2 detection kits. All four kits demonstrated good linear regression. The Ct values obtained for the two target genes were highly correlated, with correlation coefficients greater than 0.99. Among the three RNA extraction-free kits, SX08 exhibited the highest R² and r-values.
The SS kit had the lowest CV, indicating the most stable results. The SS kit utilized magnetic beads for nucleic acid extraction, and previous reports have highlighted that this method provides higher stability compared to methods without nucleic acid extraction (18). The KYD had the highest CV, indicating greater variability. Among the three RNA extraction-free kits, SX08 showed the lowest CV and the highest stability for Z0. The SX002 and SX08 demonstrated the highest accuracy in detecting the N and ORF1ab genes, respectively. The SX08 also had the lowest LOD among the RNA extraction-free kits.
The KYD kit failed to detect the ORF1ab gene, likely due to an interaction gap in its primer/probe design that hindered the detection of the target region covered by the pseudovirus particles used in the Z0 sample. In the analysis of clinical samples, the qualitative results of the SS and SX08 kits showed 100% agreement with the actual results. The KYD exhibited strong detection ability for samples with high viral loads but weaker detection ability for samples with low viral loads. In the detection of the Omicron variant, all kits tested positive for both target genes, except for KYD, which only tested positive for the ORF1 gene of the BA.5 variant at 7.5 × 102 copies/mL.
The SARS-CoV-2 genome has undergone evolutionary mutations since the COVID-19 outbreak. Although these RNA extraction-free SARS-CoV-2 detection kits were in clinical use before the emergence of the Omicron variant, they were still able to detect different Omicron variants, indicating a high degree of conservation in the primer/probe regions recognized by these kits. However, some RNA extraction-free SARS-CoV-2 nucleic acid detection kits may not efficiently detect low virus concentration samples of the Omicron variants. Similar results have been reported in other studies. Visseaux et al. (17) found that different extraction-free SARS-CoV-2 RT-PCR assays exhibited lower sensitivity for low viral load samples. Morecchiato et al. (12) also reported a loss of accuracy for extraction-free protocols in samples with low viral loads, leading to false-negative results in cases where conventional tests yielded high Ct values.
Multiple SARS-CoV-2 detection kits demonstrate good agreement in terms of qualitative results, although their Ct values differ significantly (19, 20). The present study indicates that although the RNA extraction-free nucleic acid test kits and the magnetic bead extraction nucleic acid test kit yielded consistent qualitative results, their Ct values varied. Specifically, the KYD test exhibited significantly lower Ct values compared to the other kits. The KYD test is primarily designed for rapid screening, featuring a shortened detection time of only 30 minutes with a rapid reaction program. Although the program includes 45 cycles, the first 15 cycles are pre-amplification cycles during which no fluorescence signals are collected.
The Ct values serve as a valuable parameter for the semi-quantitative assessment of viral load. However, the use of Ct values in the management of COVID-19 remains controversial due to variations across different test systems (21). RNA extraction maximizes the delivery of RNA from clinical samples into the RT-PCR reaction system while minimizing potential “interference” that may negatively affect the performance of the PCR reaction.
The main disadvantages of the three RNA extraction-free kits lie in the lower concentration and purity of the RNA samples. Fewer RNA copies are added to the reaction, and potential PCR interferences remain. Consequently, the overall detection performance is inferior to that of the magnetic bead extraction method. However, RNA extraction-free kits are easy to use, have shorter detection times, lower economic costs, and provide reliable qualitative results, making them convenient and practical.
Nonetheless, large Ct values and single-gene amplification results should be carefully considered in clinical applications to avoid false-negative results. For RNA extraction-free nucleic acid detection kits, further optimization of RNA concentration is crucial to improve detection accuracy. Additionally, continuous monitoring of the SARS-CoV-2 genome sequence and timely optimization of detection kits are essential to ensure that the primer and probe sequences in different RT-PCR detection systems adequately cover the target genes.
5.1. Conclusions
Many laboratories have chosen to utilize multiple nucleic acid detection methods, including nucleic acid extraction and RNA extraction-free approaches. However, testing systems may exhibit variations in performance indicators due to differences in testing principles and techniques. Therefore, laboratories should comprehensively consider factors such as specimen source, instruments, reaction detection systems, and applicable scenarios when selecting the appropriate detection kit. Although Ct values obtained from SARS-CoV-2 nucleic acid detection are valuable for clinical diagnosis and treatment, they can vary across different assays. Hence, variations in the performance indicators of assay kits should be carefully considered when making clinical decisions based on Ct values.