1. Background
Staphylococcus aureus is a Gram-positive coccus commonly found on the skin and mucous membranes. This bacterium is a potent pathogen, causing both hospital-acquired and community-acquired infections, such as endocarditis, pneumonia, toxic shock syndrome, abscesses, and more (1, 2). Methicillin-resistant S. aureus strains (MRSA) are a serious concern in hospital infections, leading to significant complications in the recovery of patients (3). Resistance to methicillin in MRSA strains occurs through the production of a specific binding protein, penicillin-binding protein 2a (PBP2a), which binds very weakly to beta-lactam antibiotics. Penicillin-binding protein 2a is encoded by the mecA gene, which is transmitted via a large gene chromosomal cassette [Staphylococcal cassette chromosome mecA (SCCmecA)] located in the chromosomes of resistant strains (4, 5).
Strains containing this gene exhibit resistance to various other antibiotics, including tetracycline, macrolides, and aminoglycosides, which not only complicate treatment but also promote colonization and spread within the hospital environment and to other patients (6). Methicillin-resistant S. aureus strains are generally categorized into two types: (1) Hospital-acquired MRSA (HA-MRSA), and (2) community-acquired MRSA (CA-MRSA). These two types can be distinguished by SCCmec types. Staphylococcal cassette chromosome mec types I to III are typically associated with HA-MRSA strains, while SCCmec types IV and V are primarily found in CA-MRSA infections. Recently, the prevalence of CA-MRSA strains has increased, significantly altering the global epidemiology of MRSA (7).
According to previous studies, patients infected with MRSA tend to have longer hospital stays compared to those infected with methicillin-susceptible S. aureus (MSSA). As a result, the treatment costs are higher, and there is an increased risk of infection progressing to bacteremia and higher mortality rates. Therefore, the rapid diagnosis of MRSA and the appropriate treatment of this bacterium are critical in preventing the spread of infection (8, 9). In the central region of Iran, limited data is available on the prevalence of MRSA, its resistance patterns, and SCCmec types, creating a significant knowledge gap. This study aims to characterize MRSA isolates from this region, providing insights that can inform local infection control measures, enhance antibiotic stewardship, and address the rising prevalence of CA-MRSA in Iranian hospitals.
2. Objectives
This study aims to investigate the frequency of MRSA strains, determine their antibiotic resistance pattern using the disk diffusion method, and evaluate the panton-valentine leukocidin (PVL) and SCCmec types in MRSA isolates obtained from different clinical specimens in Yazd, Iran.
3. Methods
In this descriptive cross-sectional study conducted from November 2021 to June 2022, a total of 230 cases were collected using a convenience sampling method from various clinical samples (such as blood, lung secretion, urine, wound, and others) at 6 therapeutic and diagnostic centers affiliated with Yazd University of Medical Sciences, Yazd, Iran (Shahid Sadoughi, Shahid Rahnamon, Afshar, Shohadaye Kargar, Shohadaye Mehrab hospitals, and the Central Laboratory).
3.1. Bacterial Isolates Identification
Specimens collected from patients for the isolation of S. aureus were cultured on blood agar medium (Liofilchem-Italy) and incubated at 37°C for 24 hours. Golden or white colonies, composed of gram-positive cocci arranged in clusters, were identified using standard biochemical tests, including catalase and coagulase production, evaluated by the tube method with rabbit-citrated plasma. Furthermore, the colonies were cultivated on mannitol-salt agar (Liofilchem-Italy) and DNase media (Liofilchem-Italy) for further analysis (10). Staphylococcus aureus isolates were cultured in tryptic soy broth (TSB) containing 20% sterile glycerol and stored at -70°C.
3.2. Antimicrobial Susceptibility Test
Antibiotic sensitivity testing was performed using the disc diffusion method (Kirby-Bauer) according to CLSI 2019 guidelines on Mueller-Hinton agar medium (Liofilchem, Italy) (10). The antibiotic discs used (Mast, England) included: Gentamicin (10 µg), erythromycin (15 µg), tetracycline (30 µg), rifampin (5 µg), clindamycin (2 µg), ciprofloxacin (5 µg), trimethoprim-sulfamethoxazole (23.75/1.25 µg), and linezolid (30 µg). The sensitivity to vancomycin was determined using the E-test method to measure the minimum inhibitory concentration (MIC) (11). Staphylococcus aureus ATCC 25923 and MRSA ATCC 43300 were used as control isolates.
3.3. Identification of Methicillin-Resistant Staphylococcus aureus Isolates
Methicillin-resistant S. aureus isolates were identified using cefoxitin (FOX, 30 µg, Mast, England) according to the CLSI 2019 guidelines (11).
3.4. DNA Extraction and Confirmation of Staphylococcus aureus Isolates
The boiling method was used to extract the bacterial genome, as described previously (12). PCR was then performed to detect the femA gene and PVL. The oligonucleotide primers used in this study are shown in Table 1. PCR reactions were carried out in a total volume of 20 microliters, which included PCR master mix (Ampliqon, Denmark), primers at a final concentration of 4 picomoles, and bacterial genome at a concentration of 100 ng. Sterile distilled water was added to reach a final volume of 20 microliters. The DNA thermocycler was programmed with the following temperature settings: An initial denaturation step at 94°C for 5 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 50 seconds, and extension at 72°C for 30 seconds. The program concluded with a final elongation step at 72°C for 10 minutes.
| Genes | Primer Sequence (5′-3′) | Product Size (bp) | Target | I | II | III | IV | V | References |
|---|---|---|---|---|---|---|---|---|---|
| PVL-F | AGAAGATACAAGTAGCGATAAGTG | 527 | (2) | ||||||
| PVL-R | AAGGATTGAAACCACTGTGTAC | ||||||||
| femA-F | CGATCCATATTTACCATATCA | 450 | (13) | ||||||
| femA-R | ATCACGCTCTTCGTTTAGTT | ||||||||
| B | ATTGCCTTGATAATAGCCYTCT | 937 | ccrA2-B | * | * | (14) | |||
| a3 | TAAAGGCATCAATGCACAAACACT | ||||||||
| ccrCF | CGTCTATTACAAGATGTTAAGGATAAT | 518 | ccrC | * | * | (14) | |||
| ccrCR | CCTTTATAGACTGGATTATTCAAAATAT | ||||||||
| 1272F1 | GCCACTCATAACATATGGAA | 415 | IS1272 | * | * | (14) | |||
| 1272R1 | CATCCGAGTGAAACCCAAA | ||||||||
| 5RmecA | TATACCAAACCCGACAACTAC | 359 | mecAIS431 | * | (14) | ||||
| 5R431 | CGGCTACAGTGATAACATCC |
3.5. Multiplex PCR Assay for Staphylococcal Cassette Chromosome mecA Detection
A multiplex PCR assay was used for SCCmec gene typing, as described by Boye et al. The primers specified in Table 1 were used to determine the SCCmec types (14). To amplify the SCCmec genes, the temperature program included an initial denaturation at 94°C for 4 minutes, followed by 30 cycles consisting of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 1 minute. The program was completed with a final elongation step at 72°C for 4 minutes. Distilled water was used as a negative control. The PCR products were then electrophoresed on a 1% agarose gel with a DNA Ladder (100 bp, Smo Bio, Denmark).
3.6. Statistical Analysis
The collected data were analyzed using SPSS version 16 software, with statistical tests such as the chi-square and Fisher's exact tests applied for the analysis.
4. Results
In this study, out of 230 S. aureus isolates, 116 (50.4%) were isolated from men, and 114 (49.6%) were isolated from women. The average age of the patients was 43.57 years, ranging from 1 to 92 years, with a standard deviation of 24.352. Additionally, 78 isolates were identified as MRSA. Of these 78 MRSA isolates, 32 (41%) were from men, and 46 (59%) were from women (P-value < 0.05). Among all isolates, 84.3% were from hospitalized patients, while 15.7% were from outpatients (P-value = 0.05). Wound samples accounted for 109 (47.4%) of the S. aureus isolates, followed by 66 (28.7%) from blood samples, 22 (9.6%) from urine samples, 15 (6.5%) from bronchi, and 18 (7.8%) from other body parts (P-value > 0.05). According to Table 2, the highest resistance was observed against erythromycin (83.3%) and tetracycline (56.4%). All samples were susceptible to linezolid, and no resistance to vancomycin was detected.
| Antibiotics | Sensitive | Intermediate | Resistant |
|---|---|---|---|
| GM | 65 (83.3) | 0 | 13 (16.7) |
| E | 5 (6.4) | 8 (10.3) | 65 (83.3) |
| TE | 33 (42.3) | 1 (1.3) | 44 (56.4) |
| RA | 64 (82.1) | 0 | 14 (17.9) |
| CC | 50 (64.1) | 6 (7.7) | 22 (28.2) |
| CP | 37 (47.4) | 8 (10.3) | 33 (42.3) |
| SXT | 54 (69.2) | 4 (5.1) | 20 (25.6) |
| LNZ | 78 (100) | 0 | 0 |
| V | 78 (100) | 0 | 0 |
Abbreviations: GM, gentamicin; E, erythromycin; TE, tetracycline; RA, rifampin; CC, clindamycin; CP, ciprofloxacin; SXT, cotrimoxazole; LNZ, linezolid; V, vancomycin.
a Values are expressed as No. (%).
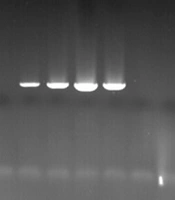
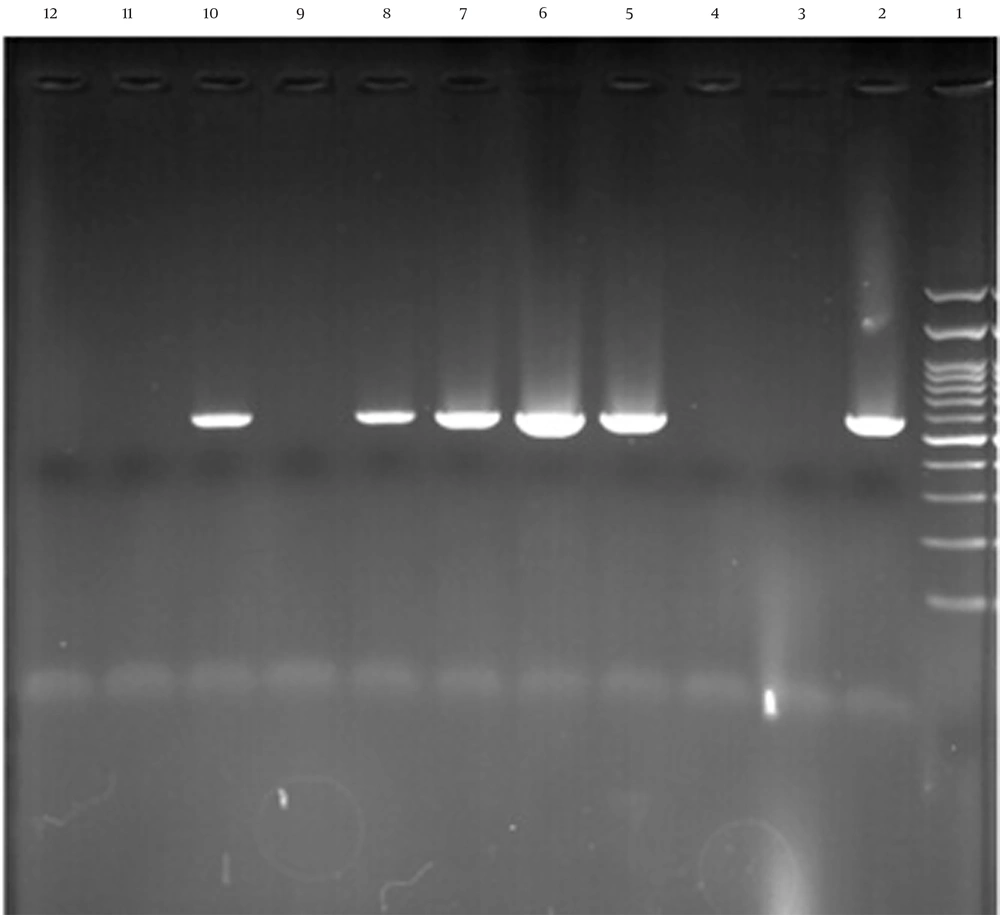
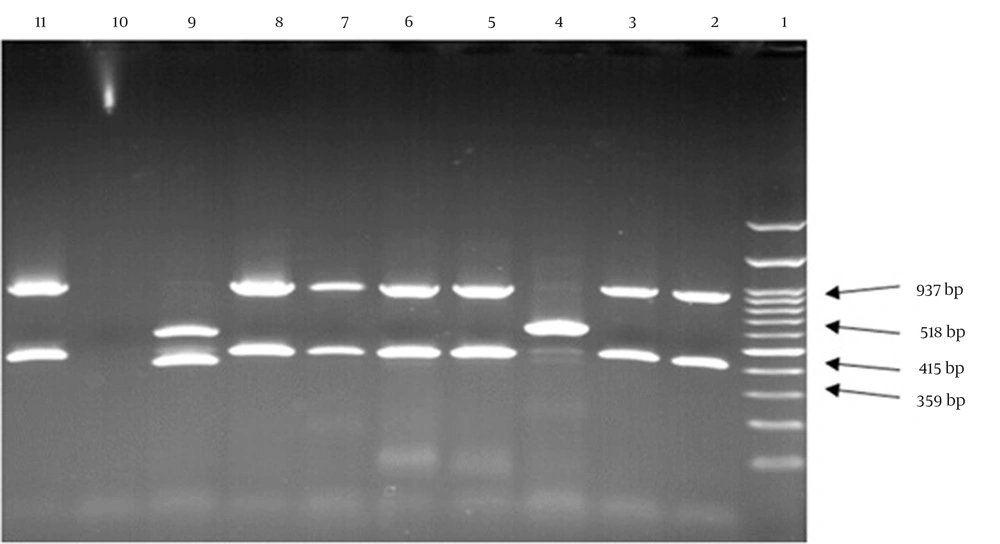
In this study, all S. aureus isolates were confirmed by the femA gene. Among the 78 MRSA isolates, 23 (29.5%) contained the PVL gene (Figure 1). The frequency of the PVL gene was higher in wound and blood samples than in other samples, with a higher prevalence in HA-MRSA compared to CA-MRSA isolates (P-value > 0.05). The frequency of PVL genes in different types of isolated samples is shown in Table 3. Furthermore, the MRSA isolates were further characterized by SCCmec gene typing (Figure 2). Among the 78 MRSA isolates, the majority (75.6%, n = 59) were classified as type IV, followed by type III (11.5%, n = 9), type V (10.3%, n = 8), and type I (2.6%, n = 2).
The results of panton-valentine leukocidin (PVL) gene amplification by agarose gel electrophoresis test, column 1 is 100 pb ladder, columns 2, 5, 6, 7, and 8 related to PVL gene, column 10 is a positive control, columns 3, 4, 9, and 11 are negative isolates and column 12 is negative control.
| Sample Type | PVL Gene | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Wound | 12 (32.4) | 25 (67.6) | 37 (100) |
| Blood | 6 (28.6) | 15 (71.4) | 21 (100) |
| Bronchus | 0 | 7 (100) | 7 (100) |
| Urine | 3 (50) | 3 (50) | 6 (100) |
| Other samples | 2 (28.6) | 5 (71.4) | 7 (100) |
| Total | 23 (29.5) | 55 (70.5) | 78 (100) |
Abbreviation: PVL, Panton-valentine leukocidin.
a Values are expressed as No. (%).
5. Discussion
Ensuring the prompt identification and treatment of MRSA infections is crucial for preventing the transmission of infection and minimizing the potential risk of patient mortality. In addition to being resistant to methicillin and beta-lactam drugs, MRSA strains exhibit resistance to several other antibiotics (12, 15). In this study, 230 S. aureus isolates were detected and evaluated using phenotypic and genotypic methods to identify MRSA strains. The samples included 109 from wounds, 66 from blood, 22 from urine, 15 from lung samples, and 18 from other sources. Methicillin-resistant S. aureus Istrains displayed varying levels of resistance to different antibiotics. Specifically, the resistance rates were as follows: Erythromycin 83.3%, tetracycline 56.4%, ciprofloxacin 42.3%, clindamycin 28.2%, cotrimoxazole 25.6%, rifampin 17.9%, and gentamicin 16.7%. However, all strains exhibited sensitivity to vancomycin and linezolid.
Similar results were reported in a study conducted in Brazil by Lima et al. on patients with underlying cystic fibrosis. Among 28 methicillin-resistant S. aureus isolates, the resistance rates to antibiotics such as cotrimoxazole, ciprofloxacin, clindamycin, erythromycin, gentamicin, and tetracycline were 14.3%, 25%, 25%, 53.3%, 35.7%, and 21.4%, respectively. All isolates were sensitive to vancomycin and linezolid (16). In another study conducted by Boswihi et al. in Kuwait, out of 1327 MRSA isolates from clinical samples, the resistance rates to erythromycin, clindamycin, ciprofloxacin, and tetracycline were 39.2%, 39.2%, 38.2%, and 32.1%, respectively, with all isolates being sensitive to vancomycin and linezolid antibiotics (17). A study by Samadi et al. in Iran on 98 MRSA isolates showed resistance rates to rifampin (12%), gentamicin (24%), trimethoprim-sulfamethoxazole (24%), clindamycin (57%), and erythromycin (62%), with all isolates being sensitive to vancomycin and linezolid (18).
In the present study, of the 230 S. aureus isolates, 78 were identified as MRSA, making the frequency of MRSA isolates 33.9%. The highest frequency was related to wound samples, which accounted for 47.4% of the MRSA isolates. Our findings align with those of Sadeghi and Mansouri in Kerman, Iran, where 56.8% of 162 S. aureus isolates were MRSA, with the majority coming from urine samples (19). In a study by Wangai et al. in Africa, the prevalence of MRSA among S. aureus isolates was 53.4%, with the highest frequency observed in pus samples (20). Similarly, a study by Wang et al. in China found that 40.6% of 32 S. aureus isolates were MRSA (21).
In our study, 23 (29.5%) of the 78 MRSA isolates tested positive for the PVL gene. Of these, 8 were found in men and 15 in women. However, no significant association was observed between gender and the prevalence of the PVL gene. The majority of PVL-positive isolates were from wound samples, with 12 isolates (52.1%). A study conducted in Iran by Hessari et al. found that the PVL gene was present in 20% of the samples. Additionally, in that study, no significant relationship was found between the presence of the PVL gene and the sample type (2). In a study conducted by Tajik et al. in 2020 among the Iranian population of Tehran, the prevalence of MRSA and PVL-positive isolates was investigated. The presence of the PVL gene was detected in 11.2% of the samples (22). In our study, the majority of PVL-positive samples were related to bronchial and wound samples, a finding that is consistent with a study conducted by Ahmad et al. in 2020 in Malaysia. Their study found that 20% of the isolates were PVL-positive (23). Furthermore, the frequency of the PVL gene was higher in the HA-MRSA samples of this study. While PVL genes were first reported in CA-MRSA strains (24), an analysis of MRSA isolates in the Netherlands in 2003 revealed the presence of MRSA strains carrying PVL genes, suggesting that these strains can be found both in the community and within the hospital environment. As noted earlier, MRSA strains are prevalent pathogens in hospitals and have also been detected in the community. Community-acquired Methicillin-resistant S. aureus strains and HA-MRSA can be distinguished by identifying their SCCmecA types (25).
In the current study, among 78 MRSA isolates, 59 (75.6%) were classified as type IV, 9 (11.5%) as type III, 8 (10.3%) as type V, and 2 (2.6%) as type I. Type II was not identified in this study. Furthermore, 20 isolates (87%) contained PVL genes, which were primarily associated with the CA-MRSA group. According to the classification, 85.9% of MRSA strains belonged to types IV and V, indicating an increase in the prevalence of CA-MRSA and a shift in its epidemiology in Yazd (Iran) hospitals, which requires special attention. Goudarzi et al. conducted a study in Iran on phenotypic and molecular characteristics of MRSA containing the PVL gene. They found that 62 (88.6%) isolates were type IV, and 8 (11.4%) were type V, with no other types identified. Their study indicated an increase in CA-MRSA isolates in hospitals (26). In another study by Peng et al. in China, 70.9% of isolates were type IV, 15.4% were type V, and 10.3% were type III (27). In Saudi Arabia, Eed et al. found that 34.3% of isolates were type I, 20% were type III, 20% were type IV, 15.8% were type V, and 10% were type II (7). Similarly, Alfouzan et al. in Kuwait reported that 39.5% of isolates were type IV, 34.4% were type III, and 25.8% were type V (28).
Our study, in line with others, clearly indicates that the prevalence of CA-MRSA has surpassed that of HA-MRSA, resulting in a shift in its epidemiology. As a consequence, there is an urgent need for further investigation into CA-MRSA. This study has some limitations, including a limited sample size that may not fully represent the central region of Iran. Additionally, while the study focuses on antibiotic resistance and SCCmecA types, it does not explore other virulence factors or genetic characteristics.
5.1. Conclusions
The present study demonstrates that MRSA isolates from the hospital environment are resistant to all antibiotics except vancomycin and linezolid. Therefore, hospital personnel should take precautions to prevent the spread of such strains within the hospital. Proper treatment of patients and stringent control of unnecessary antibiotic use are strongly recommended. Based on our study and other research on SCCmec types, it is evident that the prevalence of the CA-MRSA group is rising in both Iranian and global hospitals, surpassing the prevalence of the HA-MRSA group. Additionally, the presence of the PVL pathogenic gene in the CA-MRSA group, coupled with its high antibiotic resistance, highlights the need for periodic detection of CA-MRSA isolates and monitoring of their antibiotic resistance in hospitals.