1. Background
The Enterobacteriaceae family includes several important human pathogens, such as Escherichia coli, Klebsiella pneumoniae, and Salmonella species (1). Uropathogenic E. coli (UPEC), a specific pathotype of E. coli responsible for urinary tract infections (UTIs), encodes various adhesive and secretory virulence factors (2). Based on genetic characteristics, E. coli is classified into various phylogroups, with certain phylogroups, such as B2 and D, known to be associated with specific pathogenic E. coli strains. Among them, sequence type 131 (ST131) is a globally recognized high-risk clone causing significant extraintestinal infections (3). Antibiotic resistance in E. coli has recently become a major concern due to its ability to acquire and spread resistance genes through various mechanisms, such as β-lactamase production (4). Extended-spectrum β-lactamase (ESBL)-producing E. coli (EP-E. coli) hydrolyzes β-lactams, rendering these bacteria resistant to β-lactam antibiotics (5).
Carbapenemase-producing E. coli (CP-E. coli) can hydrolyze carbapenem antibiotics, which are often used as a last-resort treatment for multidrug-resistant (MDR) bacterial infections (6). The emergence of carbapenemase-producing UPEC strains, particularly ST131, poses a significant public health threat due to limited treatment options (7). Furthermore, E. coli can form biofilms, which reduce antibiotic penetration and facilitate the exchange of virulence and resistance genes (8). The production of ESBLs and carbapenemases indicates a high level of antibiotic resistance, forcing healthcare providers to rely on alternative antibiotics such as colistin, temocillin, and ceftazidime-avibactam (CAZ/AVI). However, data on carbapenem-resistant ST131 clones remain limited. Herein, we investigated the molecular characteristics, antibiotic resistance, and virulence factors of EP-E. coli, particularly the ST131 clone, causing UTIs in hospitalized patients.
2. Objectives
This study aimed to evaluate the genetic characteristics of ESBL- and carbapenemase-producing E. coli isolates.
3. Methods
3.1. Sample Collection
This study was conducted from January 2019 to December 2020. A total of 100 clinical E. coli isolates were collected from 300 urine samples of UTI patients hospitalized in various wards of a hospital in Tehran. The samples were inoculated into MacConkey agar and blood agar (Conda, Spain) and identified using biochemical tests (9).
3.2. Antibiotic Susceptibility Test
Susceptibility to the following antibiotics was tested using the disc diffusion method: Nitrofurantoin (NFT; 300 µg), fosfomycin (FO; 30 µg), gentamicin (GEN; 10 µg), ampicillin (AMP; 10 µg), aztreonam (ATM; 30 µg), trimethoprim/sulfamethoxazole (SXT; 25 µg), ciprofloxacin (CIP; 5 µg), nalidixic acid (NAL; 30 µg), cefotaxime (CTX; 30 µg), ceftazidime (CZA; 30 µg), imipenem (IPM; 10 µg), meropenem (MEM; 10 µg), ertapenem (ETP; 10 µg), piperacillin/tazobactam (TZP; 100/10 µg), AMP/sulbactam (SAM; 10/10 µg), amoxicillin/clavulanic acid (AMC; 20/10 µg), and amikacin (AMK; 30 µg) (Mast, UK). Klebsiella oxytoca ATCC 13182 and E. coli ATCC 25922 were used as control strains (10).
3.3. Antibiotic Resistance in Uropathogenic Escherichia coli Isolates
The minimum inhibitory concentrations (MICs) of MEM and CAZ/AVI in carbapenem-resistant E. coli isolates were determined using the E-test method (BioMérieux, France) (11). Temocillin resistance was evaluated using the disk diffusion method with a temocillin disk (30 μg; Liofilchem, Italy). The MIC of colistin (Sigma, USA) was measured using the broth microdilution method with colistin sulfate powder (19,000 IU/mg) in 96-well round-bottom microtiter plates (MTPs). Escherichia coli ATCC 25922 and Proteus mirabilis ATCC 12453 were used as control strains.
3.4. Extended-Spectrum β-Lactamase and Carbapenemase Production Determination
Identification of ESBL-producing E. coli (EP-E. coli) isolates was determined by initial ESBL screening using the Kirby-Bauer disk diffusion method with CTX and CZA (30 µg each). The ESBL confirmatory test was conducted using the combination disk test recommended by the CLSI, which included CTX/CTX + clavulanic acid and CZA/CZA + clavulanic acid. Klebsiella pneumoniae ATCC 700603 and E. coli ATCC 25922 were used as control strains.
Carbapenemase-producing isolates were identified based on resistance to ETP, IPM, and MEM, with a MIC ≥ 4 µg/mL for MEM. Confirmation of carbapenemase production was performed using the combined disk test, which involved a reduction in the zone diameter in the presence of EDTA in the MEM/MEM ± EDTA (0.5 M) combination disk test. Additionally, the Carba NP test was conducted by observing a color change in phenol red with ZnSO₄ and an IPM-cilastatin tube, following CLSI guidelines (11).
3.5. Biofilm Formation Assay
Biofilm production was assessed phenotypically using the MTP assay, as described in previous studies (12, 13). Briefly, an initial culture was prepared in trypticase soy broth (Merck, USA) containing 1% glucose and incubated at 37°C for 24 hours. The wells of a 96-well plate were then washed, and precipitates were fixed using methanol before being stained with 10% crystal violet for 15 minutes. Ethanol was subsequently added, and absorbance was measured at a wavelength of 590 nm using an ELISA reader. The level of biofilm formation was determined based on the cut-off optical density (ODc) and the OD of the sample isolate, following previously established criteria (12-14).
3.6. Amplification of Genes by Polymerase Chain Reaction
DNA was extracted using the boiling method (15). Polymerase chain reaction (PCR) was performed to amplify resistance genes (blaCTX-M, blaTEM, blaSHV, blaNDM, blaVIM, blaIMP, blaKPC, and blaOXA-48), virulence genes (piccsgA, iutA, ibeA, vat, hlyA, sat, traT, cdt, cnf1, kpsMTII, and tcpC), serogroups (O1, O2, O4, O6, O7, O12, O15, O16, O18, O25, O75, and O157), and phylogroups (TspE4.C2, chuA, and yjaA), using specific primers, some of which are listed in Table 1 (16, 17).
| Target Genes and Primers | Sequence (5´- 3´) | Amplicon Size (bp) | Annealing Temp (ºC) | Reference |
|---|---|---|---|---|
| pabB | 347 | 63 | (18) | |
| F | TCCAGCAGGTGCTGGATCGT | |||
| R | GCGAAATTTTTCGCCGTACTGT | |||
| trpA | 427 | 63 | ||
| F | GCTACGAATCTCTGTTTGCC | |||
| R | GCAACGCGGCCTGGCGGAAG | |||
| rfbO25b | 300 | 60 | ||
| F | ATACCGACGACGCCGATCTG | |||
| R | TGCTATTCATTATGCGCAGC | |||
| blaCTX-M | 569 | 55 | (19) | |
| F | CGCTGTTGTTAGGAAGTGTG | |||
| R | GGCTGGGTGAAGTAAGTGAC | |||
| blaKPC | 452 | 56 | (20) | |
| F | ATCTGACAACAGGCATGACG | |||
| R | ACGGCCAACACAATAGGTG | |||
| blaNDM | 203 | 56 | ||
| F | GCAGGTTGATCTCCTGCTTG | |||
| R | ACGGTTTGGCGATCTGG | |||
| blaOXA-48 | 438 | 56 | ||
| F | GCAGGTTGATCTCCTGCTTG | |||
| R | ATCAAGTTCAACCCAACCG | |||
| mcr-1 | 309 | 55 | (21) | |
| F | CGGTCAGTCCGTTTGTTC | |||
| R | CTTGGTCGGTCTGTAGGG |
Abbreviations: bp, base pair; F, forward; R, reverse; ST131, sequence type 131.
3.7. Genetic Relatedness of the Isolates
The chromosomal DNA was digested using the XbaI enzyme (22). The DNA of the Salmonella serotype Braenderup strain H8912 was used as a molecular weight standard. The dendrogram was constructed using Gel Compare II. Isolates with a Dice Similarity Index ≥ 80% were considered to belong to the same pulsed-field gel electrophoresis (PFGE) cluster.
3.8. Molecular Characterization of Sequence Type 131 Clone
The ST131 clones were identified by PCR of ST131-specific single nucleotide polymorphisms in the mdh and gyrB genes and confirmed by multi-locus sequence typing (MLST) (23).
3.9. Statistical Analysis
Statistical analysis was performed using R software version 3.3.3. Data were interpreted based on frequency distribution and percentage. A P-value ≤ 0.05 (95% confidence interval) was considered statistically significant.
4. Results
4.1. Antimicrobial Resistance Patterns of Isolates
Among the 300 urine samples included in this study, 100 E. coli isolates were detected. The demographic information and distribution of E. coli isolates are presented in Table 2. The isolates exhibited resistance patterns to the following antibiotics: Ampicillin, 92% (n = 92); CTX, 85% (n = 85); ceftazidime-avibactam, 74% (n = 74); trimethoprim-sulfamethoxazole (SXT), 63% (n = 63); ATM, 54% (n = 54); NAL, 51% (n = 51); CIP, 49% (n = 49); ampicillin-sulbactam (SAM), 38% (n = 38); amoxicillin-clavulanate (AMC), 31% (n = 31); piperacillin-tazobactam (TZP), 26% (n = 26); nitrofurantoin (NFT), 21% (n = 21); AMK, 20% (n = 20); gentamicin (GEN), 18% (n = 18); fosfomycin (FO), 18% (n = 18); ETP, 12% (n = 12); IPM, 12% (n = 12); and MEM, 11% (n = 11).
| Characteristics | Total (N = 100) | ESBL (N = 36) | Non-ESBL (N = 64) | P-Value b |
|---|---|---|---|---|
| Gender | 0.907 | |||
| Male | 52 | 19 (52.8) | 33 (51.5) | |
| Female | 48 | 17 (47.2) | 31 (48.5) | |
| Age | 0.315 | |||
| < 40 | 18 | 5 (13.8) | 13 (20.3) | |
| 40 - 60 | 31 | 9 (25.0) | 22 (34.3) | |
| > 60 | 51 | 22 (6.1) | 29 (45.3) | |
| Clinical distribution | 0.933 | |||
| Nephrology | 40 | 14 (38.8) | 26 (40.6) | |
| Hematology | 26 | 9 (25.0) | 17 (26.5) | |
| ICU | 18 | 8 (22.2) | 10 (15.6) | |
| Emergency | 12 | 4 (11.11) | 8 (12.5) | |
| Other | 4 | 1 (2.7) | 3 (4.6) | |
| Prior antibiotic use | 0.01 | |||
| Positive | 61 | 32 (88.8) | 29 (45.3) | |
| Negative | 39 | 4 (11.1) | 35 (54.6) | |
| Prior hospitalization | 0.02 | |||
| Positive | 63 | 28 (75.6) | 35 (55.5) | |
| Negative | 37 | 9 (33.3) | 28 (44.4) | |
| Underlying disease | 0.937 | |||
| Diabetes | 27 | 10 (27.7) | 17 (26.5) | |
| Cancer | 20 | 8 (22.2) | 12 (18.7) | |
| Kidney disease | 13 | 5 (13.8) | 8 (12.5 ) | |
| Liver disease | 12 | 5 (13.8) | 7 (10.9) | |
| Hart disease | 8 | 3 (8.3) | 5 (15.6) | |
| Other | 4 | 0 (0.0) | 4 (6.25 ) | |
| Non Underlying disease | 16 | 5 (13.8) | 11 (17.1) | |
| Biofilm formation | 26 | 14 (38.8) | 12 (17.2) | 0.04 |
Abbreviation: ESBL, extended-spectrum β-lactamase; ICU, intensive care unit.
a Values are expressed as No. (%).
b P ≤ 0.05 was considered as statistically significant.
A total of 36% (n = 36/100) of E. coli isolates were phenotypically ESBL producers. Among the ESBL-producing isolates, 38.8% (n = 14/36) and 33.3% (n = 12/36) were resistant to temocillin and carbapenems, respectively. Additionally, 30.5% (n = 11/36) were classified as CP-E. coli, with a MIC ≥ 4 µg/mL against MEM. Furthermore, 25% (n = 9/36) and 16.6% (n = 6/36) of the isolates were resistant to CAZ/AVI with an MIC ≥ 256 µg/mL and colistin with an MIC > 4 µg/mL, respectively.
The E. coli isolates demonstrated multiple resistance to cephalosporins, sulfonamides, and fluoroquinolones, with 48% (n = 48/100) classified as MDR. Prior antibiotic consumption and hospitalization were significant risk factors for the isolation of ESBL-producing E. coli isolates (P = 0.01 and P = 0.02, respectively). Notably, there was no association between age, gender, different wards, and underlying diseases with the isolation of ESBL-producing E. coli (P > 0.05).
4.2. Prevalence of Extended-Spectrum β-Lactamase and Carbapenemase Genes
Among the 36 ESBL-producing E. coli isolates surveyed, 80.5% (n = 29), 52.7% (n = 19), and 47.2% (n = 17) harbored the blaCTX-M, blaTEM, and blaSHV genes, respectively. All 100% (n = 11) of the CP-E. coli isolates carried the blaCTX-M gene and were ESBL producers. Among the 11 CP-E. coli isolates, 54.5% (n = 6) and 18% (n = 2) harbored the blaNDM and blaOXA-48 genes, respectively. Additionally, 27.2% (n = 3) of the isolates harbored both the blaNDM and blaOXA-48 genes. None of the isolates carried the blaKPC, blaIMP, and blaVIM genes.
4.3. Analysis of Isolates by Pulsed-Field Gel Electrophoresis
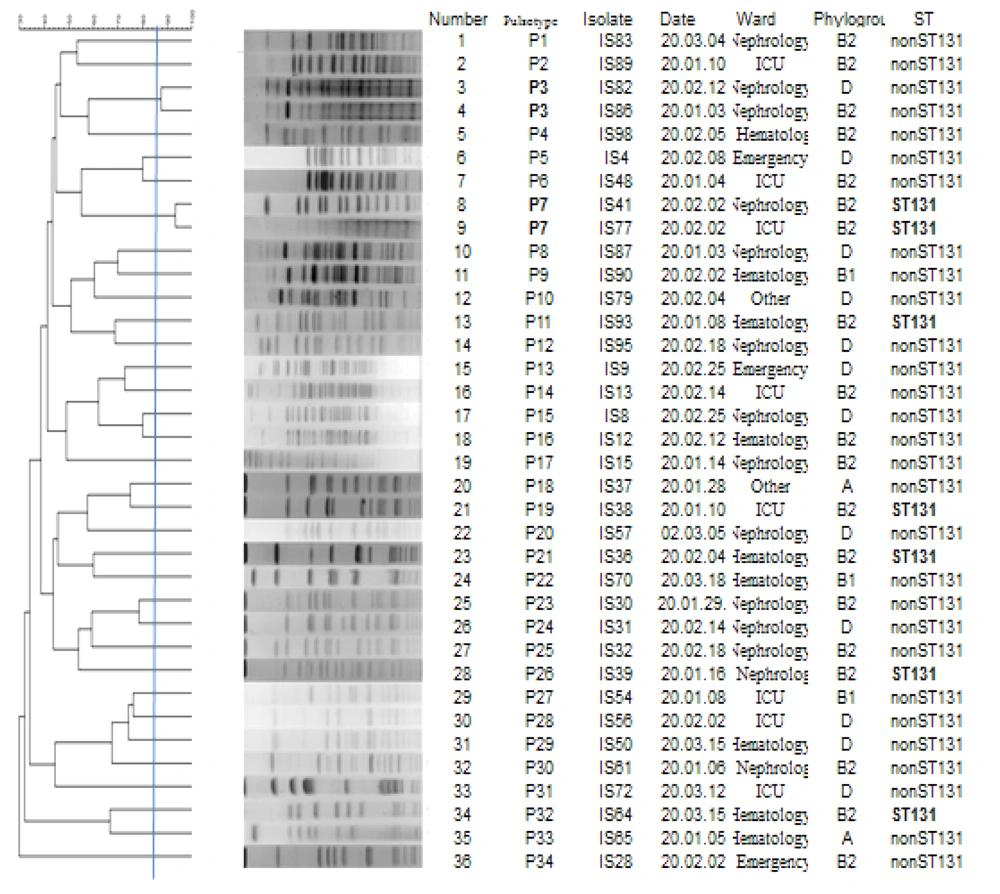
Pulsed-field gel electrophoresis analysis of the 36 extended-spectrum beta-lactamase-producing E. coli (EP-E. coli) isolates is illustrated in Figure 1. The analysis demonstrated 34 pulsotypes, numbered from P1 to P34, which were classified into 32 singletons and 2 clusters, each consisting of two strains.
4.4. Results of Biofilm Formation
Overall, 26% (n = 26/100) of the isolates were biofilm producers. As represented in Table 2, biofilm formation had a significant association with EP-E. coli isolates (P = 0.04). Among the EP-E. coli isolates, 14 were biofilm producers, with 28.5% (n = 4/14) being strong biofilm producers. However, 57.1% (n = 8/14) and 14.2% (n = 2/14) were moderate and weak biofilm producers, respectively.
4.5. Recognition of Phylogroups and Serogroups
The majority of the EP-E. coli isolates belonged to the phylogenetic groups B2 (52.7%; n = 19/36) and D (33.3%; n = 12/36), followed by groups B1 (8.3%; n = 3/36), A (2.7%; n = 1/36), and F (2.7%; n = 1/36). Additionally, serogroup O1 was detected in 36.1% (n = 13/36) of the isolates, followed by serogroups O25 (22.2%; n = 8/36), O75 (13.8%; n = 5/36), O18 (8.3%; n = 3/36), O15 (5.5%; n = 2/36), O4 (2.7%; n = 1/36), and O16 (2.7%; n = 1/36). The serogroup of 8.3% (n = 3/36) of the isolates could not be detected.
4.6. Identification of Virulence Factor Genes
Among the EP-E. coli, 97.2% (n = 35), 86.1% (n = 31), 83.3% (n = 30), 80.5% (n = 29), 33.3% (n = 12), 30.5% (n = 11), and 27.7% (n = 10) carried the fimH, iutA, fyuA, inh, traT, papП, and csgA genes, respectively. In addition, 30.5% (n = 11), 27.7% (n = 10), 25% (n = 9), 19.4% (n = 7), 16.6% (n = 6), 13.8% (n = 5), 13.8% (n = 5), and 8.33% (n = 3) carried the fimA, ompT, usp, sfa/foc, hly, cnf-1, afa, and iroN genes, respectively.
4.7. Detection of Sequence Type 131Clone
A total of 19.4% (n = 7/36) of the EP-E. coli isolates were identified as ST131. All the isolates (100%; n = 7/7) were detected as the O25b-ST131 clone, with 57.1% (n = 4/7) being carbapenemase-producing isolates containing the blaCTX-M, blaNDM, and blaOXA-48 genes (Table 3). Most of the carbapenemase-producing E. coli isolates belonged to serogroup O1, and three of these isolates containing the blaNDM and blaOXA-48 genes belonged to O25 (ST131 clone).
| ESBL | MEMMIC (µg/mL) | CTX MIC (µg/mL) | CAZ/AVI MIC (µg/mL) | ColistinMIC (µg/mL) | Temocillin Sensitivity | Resistance Genes | Phyl/Sero | Sequence Typing |
|---|---|---|---|---|---|---|---|---|
| Yes | 64 | 32 | 256 | 0.5 | R | CTX-M/OXA-48/NDM | B2/O25 | ST131 |
| Yes | 16 | 32 | 256 | 16 | R | CTX-M/NDM | B1/O1 | Non-ST131 |
| Yes | 8 | 32 | 256 | 32 | R | CTX-M/NDM | B2/O1 | Non-ST131 |
| Yes | 8 | 32 | 256 | 0.5 | S | CTX-M/NDM | B2/O25 | ST131 |
| Yes | 8 | 32 | 256 | 1 | R | CX-M/OXA-48/NDM | B2/O25 | ST131 |
| Yes | 128 | 32 | 256 | 16 | R | CTX-M/NDM | B2/O1 | Non-ST131 |
| Yes | 4 | 32 | 0.1 | 0.5 | R | CTX-M/OXA-48 | B2/O1 | Non-ST131 |
| Yes | 128 | 32 | 256 | 0.5 | R | CTX-M/OXA-48/NDM | B2/O25 | ST131 |
| Yes | 4 | 32 | 0.1 | 1 | S | CTX-M/OXA-48 | B1/O1 | Non-ST131 |
| Yes | 4 | 32 | 256 | 32 | R | CTX-M /NDM | B2/O1 | Non ST131 |
| Yes | 4 | 32 | 256 | 0.5 | R | CTX-M/NDM | B2/O16 | Non-ST131 |
Abbreviations: ESBL, extended-spectrum β-lactamase; CAZ/AVI, ceftazidime/avibactam; ST131, sequence type 131.
5. Discussion
Escherichia coli is one of the most common causative agents of UTIs worldwide, and certain strains of E. coli, owing to attributes such as high virulence factors and significant antibiotic resistance, are rapidly spreading globally, like the ST131 clone (2). This study found a high prevalence of MDR E. coli and observed that the antibiotic resistance patterns of these isolates were similar to those reported in other studies conducted in different clinical settings (24). Prior antibiotic consumption and hospitalization were significant risk factors for the isolation of MDR E. coli isolates. Several investigations have reported a connection between previous antibiotic use and the isolation of MDR strains (25, 26).
Temocillin and carbapenems are two options for treating EP-E. coli. temocillin is stable against ESBLs and AmpC β-lactamase and is effective in the treatment of UTI infections (27). In our study, temocillin was active against 61.2% of ESBL-producing isolates; therefore, the susceptibility of the isolates to this antibiotic can be considered an alternative treatment for such complex infections. Carbapenems are typically used to treat complicated bacterial infections with EP-E. coli isolates, and the percentage of resistance to these antibiotics varies across studies and has been rapidly increasing, particularly in developing countries, due to the excessive use of this class of antibiotics (28). The percentage of resistance to carbapenems in our study was high (30.6%), similar to some developing countries (29, 30).
Colistin is often used to treat infections caused by carbapenem-resistant isolates (31). In this study, the majority of carbapenem-resistant E. coli isolates were resistant to most available antibiotics; therefore, in some cases, colistin is often used for treating infections caused by these isolates. The prevalence rate of colistin resistance varies in different countries, with the highest rate (19%) found in Thailand and the lowest rate (0.8%) observed in South Korea (32-34). Colistin resistance in our study (16.6%) indicated the high use of this antibiotic in the treatment of carbapenem-resistant isolates in Iran due to limited new antibiotic options.
Ceftazidime-avibactam is recognized as a global new treatment alternative for carbapenem-resistant infections (35). Although this antibiotic is not approved in our country, its resistance has been recognized. Resistance to CAZ/AVI in carbapenem-resistant isolates has increased to 71.4% in countries where CAZ/AVI treatment is available, but the high rate (25%) of CAZ/AVI resistance in our study suggests that the emergence of its resistance is not related to previous CAZ/AVI treatment (36). Based on the PFGE pattern in the present study, similar genotypes were isolated from hospital wards on different dates, indicating that some resistant strains have a common origin that can disseminate across hospital wards. Therefore, the hospital infection control committee is required to identify the origin of these resistant isolates and employ effective health strategies to decrease the spread of resistant bacteria in the hospital (37).
As emphasized in studies, the intensive care unit (ICU), where the ST131 clone with a similar pattern was collected, is a major ward in disseminating resistant bacterial strains because patients are hospitalized in this ward for a long time, and they can be a source of infection. Hence, the hospital infection control committee must pay more attention to controlling the dissemination of infection in hospitals via patients, food, water, doctors, staff, and beds by surveillance and finding the source of infection. The prevalence of ESBL genes can vary depending on geographical locations, healthcare settings, and the population being studied (38). The ESBL enzymes, which hydrolyze cephalosporins (CTX, CZA, ceftriaxone, cefuroxime, and cefepime) and monobactams (ATM), are becoming a major challenge for the treatment of pathogenic bacteria (5). However, similar to a previous study conducted in our country, the prevalence of blaCTX-M is high and noticeable (39).
Carbapenemase genes are responsible for encoding enzymes that can break down and inactivate carbapenem antibiotics, which are considered last-resort antibiotics for treating severe bacterial infections. The prevalence of carbapenemase genes among carbapenem-resistant bacteria is influenced by factors such as antibiotic use, infection control practices, and the dissemination of resistant strains (40). In some parts of the world, the prevalence of carbapenemase genes can be relatively high, particularly in countries with high rates of antibiotic use and inadequate infection control measures. For instance, certain countries in Southeast Asia, the Middle East, and regions of Europe have reported high rates of bacteria producing carbapenemase (28, 41). It is worth mentioning that surveillance data on the prevalence of carbapenemase genes can vary over time and across different studies (30, 42). Local and regional surveillance programs, as well as molecular testing methods, are crucial for monitoring the prevalence and spread of carbapenemase genes.
In our study, the most frequent carbapenemase gene was blaNDM, which has been shown to cause infections with a high mortality rate (43). The biofilm formation in E. coli isolates allows bacteria to survive, persist, and cause infections. Based on available evidence, the global prevalence rate of biofilm formation varies, ranging between 56% and 100% (44). This observation indicates that various factors, including different geographical areas, low-level hygiene, and varying methods, can affect biofilm formation (44). In our study, similar to other surveys (45, 46), there was an association between biofilm formation and antibiotic resistance (P = 0.04), which could arise from antibiotic misuse and its administration without prescription in our country.
In our study, contrary to Rasoulinasab et al.'s study (47), fimH and iutA were the predominant virulence factors, while iroN was the least prevalent. This variation in gene prevalence rates may stem from the diverse sources of the samples. Similar to the review article in our country, which demonstrated that B2 and D phylogroups are predominant, in this study, B2 was the predominant phylogroup (48). It has been reported that the prevalence rate of phylogroups varies in the phylogroup pattern of E. coli, which could be ascribed to the source of isolates (49). However, the high prevalence rate of phylogroup B2 in our study was noticeable.
One of the important sequence types with high antibiotic resistance in EP-E. coli isolates is the ST131 clone, a causative agent of UTIs. There are different reported rates of this clone worldwide, which is probably due to varying times of studies conducted, geographical locations, and sample types (50-52). In our study, the isolation of blaOXA-48/blaNDM-carrying ST131 isolates is a warning of the potential for increased dissemination of carbapenem resistance genes in our country and globally.
5.1. Conclusions
Our findings demonstrated a high prevalence of virulence genes and antibiotic resistance in E. coli, which has been transferred between hospitalized patients. In the present study, CP-E. coli was found to carry blaNDM and blaOXA-48 genes belonging to ST131 O25/B2 with high antibiotic resistance, posing a risk for treatment and dissemination of resistant genes in a hospital. Understanding the characteristics of CP-E. coli in the hospital and community over different years with regard to antibiotic resistance and virulence, through rapid molecular detection and phylogenetic monitoring of such strains, can be helpful in limiting the dissemination of antibiotic resistance in the hospital.