1. Background
Malaria is one of the leading infectious diseases, with an estimated 2.1 billion cases and 11.7 million deaths between 2000 and 2022 (WHO report, 2023). Malaria control has shown promising results, and the goal of malaria elimination is gaining increasing attention. The management and elimination of malaria depend entirely on the rapid and accurate detection of patients. However, diagnosing the disease remains a significant challenge due to the variable effectiveness of detection methods across different laboratories. Conventional diagnostic methods, such as rapid diagnostic tests (RDTs) and light microscopy examination of Giemsa-stained thin and/or thick smears, are widely employed in regions where malaria is prevalent. However, microscopy can be ineffective in detecting submicroscopic and low-density malaria infections (LDMI) (1-4). As a result, microscopy depends on the microscopist's ability to identify and quantify parasite density. Additionally, microscopy can be labor-intensive, requiring significant effort and energy due to its time-consuming nature and the high level of expertise and training needed. In contrast, recent advances in molecular diagnostics have proven to be more sensitive than traditional methods (5, 6).
Polymerase chain reaction-based molecular methods offer a relatively simple and sensitive approach for detecting malaria parasites, providing enhanced efficiency compared to microscopy and RDTs. The implementation of the nested PCR strategy has significantly improved the accuracy and specificity of the PCR technique. In nested PCR, two amplification steps are performed, with the product of the first reaction serving as the template for the second reaction. The second reaction includes the oligonucleotide primer that hybridizes to the sequence present in the first product (7). Despite the availability of complete genome sequences for Plasmodium falciparum and P. vivax, most PCR-based methods still rely on the 18S rRNA gene as a target for malaria diagnosis. In the present study, the choice of 18S rRNA as the target gene was based on several factors, including its high level of sequence conservation, the availability of established primer sequences for amplification, and its presence in multiple copies across all examined organisms (8).
2. Objectives
The purpose of this report is to describe and validate an accurate molecular diagnostic method utilizing the 18S rRNA gene for diagnosing five types of human malaria. This method employs a semi-nested multiplex PCR technique, which can be performed in either a two-step or one-step format.
3. Methods
3.1. Sample Size
According to the formula, a total of 36 samples are required to achieve 80% power and a 95% confidence level. For this study, 36 positive control samples and 36 negative control samples were selected, resulting in a total of 72 samples.
3.2. Blood Samples
Seventy-two blood samples were stored at -80°C, obtained from suspected cases referred to the National Malaria Laboratory at the Faculty of Public Health, Tehran University of Medical Sciences. Two expert microscopists examined the selected samples from the laboratory freezer and documented their findings in the archive. Among these samples were those related to foreign nationals and travelers detected at stations affiliated with the School of Public Health (TUMS). One notable case involved P. ovale, which was associated with a Nigerian athlete who participated in sports competitions in Bandar Abbas, Iran (9).
3.3. Microscopy
Giemsa (5%) staining and light microscopic examination were performed. The slides were evaluated by trained and experienced microscopists, who were able to identify and differentiate malaria parasites. A minimum of 200 microscopic fields were examined before concluding that the slide was free of malaria parasites.
3.4. DNA Extraction
Template DNA for the nested PCR assay was prepared using the DNeasy Blood and Tissue Kit (VIRAGEN, Iran). The extraction was performed according to the manufacturer's protocol to extract parasite DNA from 100 µL of whole blood. The purified DNA was eluted from the column using 50 µL of elution buffer and stored at -20°C.
3.5. Gene and Primers
The target sequences of the 18S rRNA gene of P. falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi were selected, and primers were designed based on these sequences. In the first step of the nested PCR, a pair of universal primers was used. In the subsequent step, four species-specific reverse primers were designed to identify P. falciparum, P. vivax, P. malariae, and P. ovale. A specific forward primer was designed for P. knowlesi, along with a universal reverse primer. The nucleotide sequences of the primers are provided in Table 1.
| Primer | Sequence | Target Species or Genus | Amplicon Size (bp) |
|---|---|---|---|
| Universal.F | GAACGGCTCATTAAAACAGT | Plasmodium | ~1000 |
| Universal.R | GACGGTATCTGATCGTCTTC | Plasmodium | ~1000 |
| P.V.R | CACTAAAAATGGACGGATCC | P. vivax | 201 |
| P.O.R | TCCAATTACAAAACCATGAAA | P. ovale | 430 |
| P.F.R | AGAAAACCTTATTTTGAACAAAGC | P. falciparum | 634 |
| P.M.R | GTATTCAAACACAGAAACTTC | P. malariae | 803 |
| P.K.F | TGAACAGGTCAAAACTATGT | P. knowlesi | 203 |
3.6. Multiplex Semi-Nested PCR
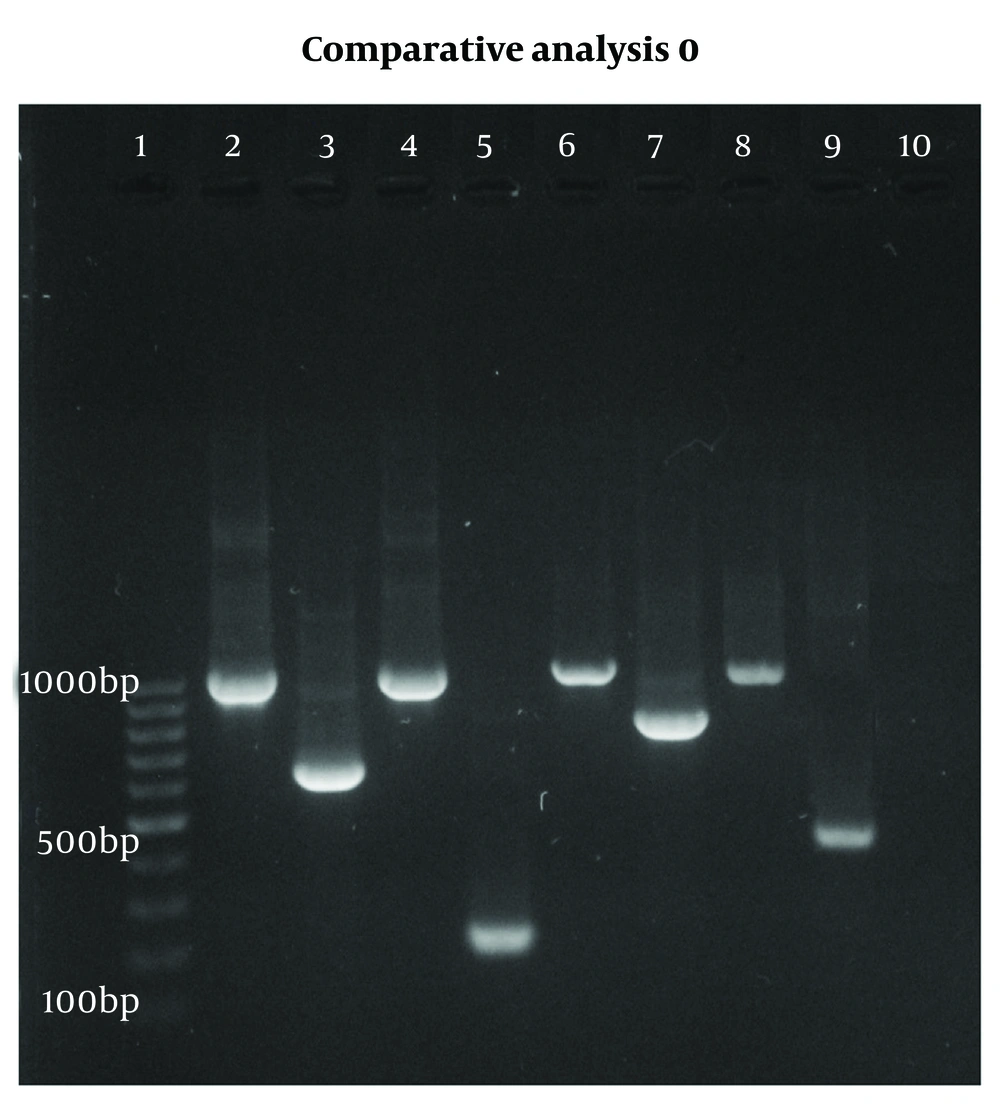
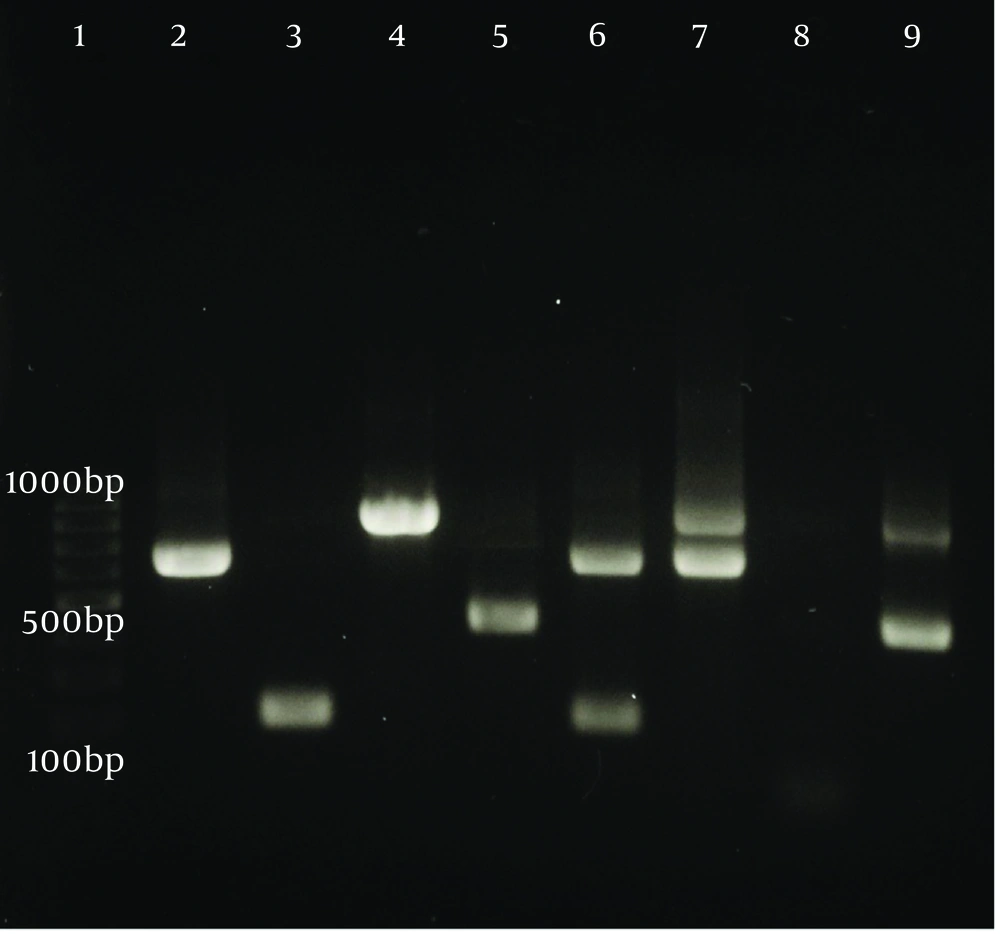
Polymerase chain reaction reactions were conducted in 25 µL volumes, which included 12 µL of master mix, 1 µL of universal forward and reverse primers, 3 µL of template DNA, and 8 µL of distilled water. The PCR assays were performed using an automated thermocycler (PTC-100TM v.7.0, MJ Research Inc., USA) with the following cycling parameters: Initial denaturation at 95°C for 5 minutes, followed by 25 cycles consisting of denaturation at 94°C for 1 minute, annealing at 58°C for 1 minute, extension at 72°C for 1 minute, and a final extension at 72°C for 5 minutes. For the second PCR cycle, the same temperature program was employed, except that the denaturation and annealing times were reduced to 30 seconds. The final PCR product was analyzed using a 1.5% agarose gel containing a safe stain, which was run at 80 V. After 45 minutes, the gel was exposed to UV light using a transilluminator (Figure 1). To confirm the ability to detect mixed contamination, PCR was performed on both confirmed mixed samples and intentionally mixed DNA samples. The resulting PCR products were then analyzed using agarose gel electrophoresis (Figure 2).
Agarose gel analysis of a polymerase chain reaction (PCR) diagnostic test for detecting Plasmodium DNA using 18S rRNA from blood samples. Lane 1: DNA marker; lanes 2, 4, 6, 8: First-stage PCR (~1000 bp); lane 3: P. falciparum (634 bp); lane 5: P. vivax (201 bp); lane 7: P. malariae (803 bp); lane 9: P. ovale (430 bp); lane 10: Negative control.
3.7. Sequencing and Phylogenetic Analysis
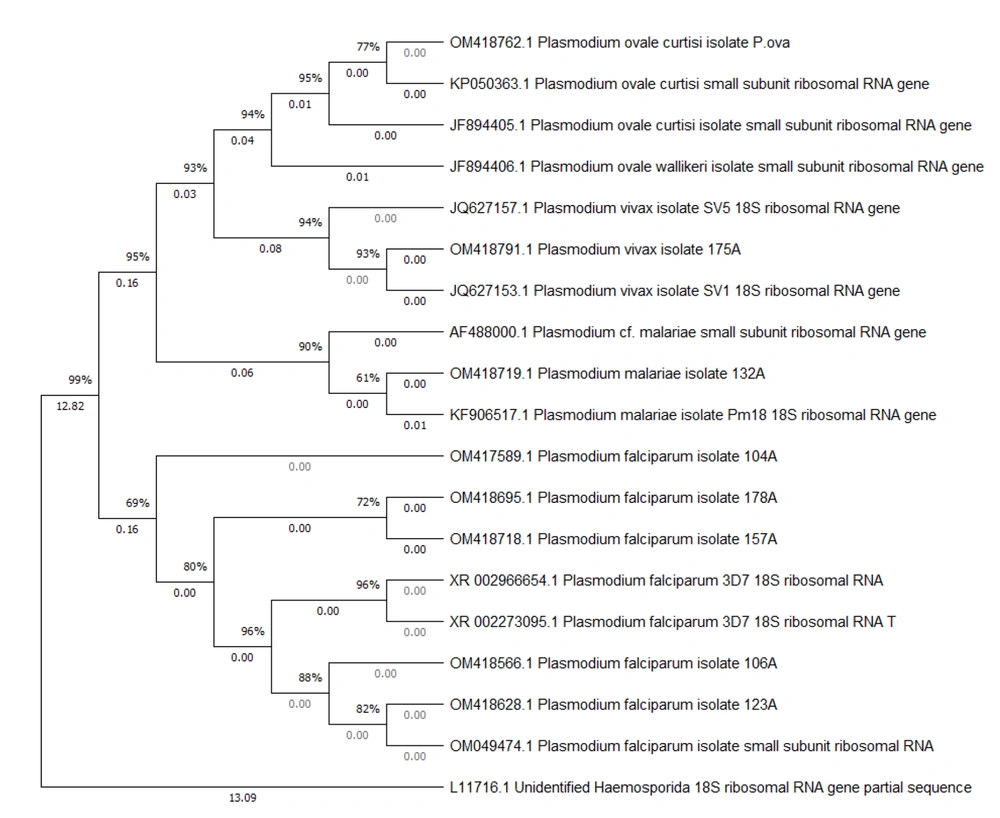
Eight Plasmodium-positive samples were selected for confirmation using molecular methods and phylogenetic analysis. Among these, one case was identified as P. ovale, one as P. malariae, one as P. vivax, and five as P. falciparum. The PCR products were sent to Pishgam Biotech Company in Iran for sequencing, and the sequence results were recorded with their respective accession numbers (OM417589.1, OM418628.1, OM418718.1, OM418719.1, OM418762.1, OM418791.1, OM418566.1, and OM418695.1) in the NCBI Gene Bank. The phylogenetic tree was constructed as follows: First, the confirmed 18S rRNA sequences for each Plasmodium species were downloaded from the NCBI database and aligned with our sequences using the ClustalW sequence alignment tool. The phylogeny was reconstructed using the maximum likelihood method based on the Tamura-Nei model, employing MEGA X version 10.2.2 software with 1 000 bootstrap iterations.
4. Results
Of the 72 samples examined, 36 were diagnosed as positive through microscopic analysis. This included 16 cases of P. falciparum, 16 cases of P. vivax, 1 case of P. ovale, 1 case of P. malariae, 1 case of mixed P. falciparum–P. vivax, and 1 case of mixed P. falciparum–P. malariae. In addition, 33 cases were diagnosed using molecular analysis, which included 16 cases of P. falciparum, 13 cases of P. vivax, 1 case of P. ovale, 1 case of P. malariae, and 2 cases of mixed P. falciparum–P. vivax, as shown in Table 2.
| Species | Method | |
|---|---|---|
| Microscopic | Semi-nested PCR | |
| Plasmodium falciparum | 16 | 16 |
| P. vivax | 16 | 13 |
| P. malariea | 1 | 1 |
| P. ovale | 1 | 1 |
| Mix (P. falciparum/P. ovale) | 1 | 2 |
| Mix (P. falciparum/P. malaria) | 1 | 0 |
| Negative | 36 | 39 |
| Total | 72 | 72 |
4.1. Discordant Results
The discordant results obtained from nested PCR compared to the microscopic method are presented in Table 3. This highlights discrepancies between the results yielded by nested PCR and those obtained through the microscopic method.
| No. | Sample Code | Nested PCR Result | Microscopy Result | Repeated Nested PCR Result |
|---|---|---|---|---|
| 1 | 123A | P. falciparum | Mix (P. falciparum/P. malaria) | P. falciparum |
| 2 | 1A | Mix (P. falciparum/P. vivax) | P. vivax | Mix (P. falciparum/P. vivax) |
| 3 | 65A | P. vivax | Mix (P. falciparum /P. vivax) | P. vivax |
| 4 | 107A | Neg | P. vivax | Neg |
| 5 | 119A | P. falciparum | P. vivax | P. falciparum |
| 6 | 36A | P. falciparum | P. vivax | P. falciparum |
| 7 | 63A | Neg | P. vivax | Neg |
4.2. Phylogenetic Analysis
The results of the phylogenetic analysis revealed two major clades and four distinct subclades. Plasmodium falciparum was placed in one clade, and the rest of the Plasmodium species were placed in another clade. P. vivax and P. ovale were placed in one subclade, and P. malariae was placed in another subclade (Figure 3). Our sample of P. ovale belonged to the P. ovale subspecies curtisi.
5. Discussion
In this study, we recorded seven cases of discrepancies between the microscopic and molecular methods. Among these, two samples of P. vivax that tested positive using the microscopic method were found to be negative when analyzed with the molecular method. This discrepancy may indicate a false positive result, or it is possible that the DNA from the samples, which were stored at -80°C, may have been damaged. In two of the samples, the species was misdiagnosed using the microscopic method. In the other two cases, a mixed infection was reported by the microscopic method, but this was not confirmed by the molecular method. Additionally, in one case, infection with the P. vivax species was diagnosed by the microscopic method, while a mixed infection with P. falciparum/P. vivax was confirmed by the molecular method. Both microscopy and RDTs as routine diagnostic methods have been shown to underestimate the prevalence of malaria parasites, particularly in asymptomatic infections with low parasitemia. Accurate identification of Plasmodium strains is critical for appropriate treatment, especially in areas endemic to multiple species. An incorrect diagnosis can lead to drug resistance and treatment failure (10, 11).
Prescribed antimalarial treatments vary based on the Plasmodium species. A delayed or missed diagnosis of P. falciparum can significantly increase the risk of developing complicated or severe malaria, which can be fatal, particularly in non-immune individuals. In many regions of the world, including Iran, P. falciparum shows inadequate response to chloroquine. Furthermore, the spread of parasite resistance to antimalarial drugs in Iran, coupled with the increasing influx of immigrants from endemic countries such as Afghanistan, complicates malaria control efforts in the country (10-12). Microscopy, traditionally regarded as the gold standard for malaria diagnosis, can detect 20 to 50 parasites per µL of blood under optimal conditions (13). However, even in regions where malaria is endemic, there are several limitations to microscopic diagnosis. These include a shortage of skilled microscopists, inadequate quality control, and the potential for misdiagnosis due to low parasitemia or mixed infections (14).
Rapid diagnostic tests, utilized as the second line in routine diagnostic protocols, can detect two proteins: HRP2 and pLDH. The cut-off levels for these tests are 1 - 50 parasites/μL for HRP2 and 51 - 100 parasites/μL for pLDH. However, RDTs are not capable of detecting all types of malaria, and there are ongoing concerns regarding their accuracy and reliability, with documented instances of both false-positive and false-negative results (15, 16). Polymerase chain reaction and PCR-based methods, such as multiplex and nested PCR, provide a sensitive and reliable alternative to conventional diagnostics for Plasmodium. In this study, we describe a specific nested PCR technique for the detection and identification of Plasmodium species. We analyzed 72 blood samples, all of which had been previously examined using routine microscopy and rapid tests. Nested PCR targeting the 18S rRNA gene has been performed with high specificity to detect a variety of organisms, including soil fungi, dermatophytes responsible for onychomycosis, Sporothrix schenckii, and protozoa such as Cryptosporidium, Theileria orientalis, Toxoplasma oocysts, Cyclospora, and Trichomonas vaginalis. This method is also employed for the diagnosis of human malaria (17-25).
In studies conducted by various researchers utilizing nested PCR, 18S rRNA gene-specific primers have been employed. However, isolating human Plasmodium species using agarose gel electrophoresis can be challenging. Our primers address this issue by producing band lengths that can be distinctly separated on high-specificity agarose gels (7, 13, 25-28). In a study conducted by Taghdiri et al. on 97 peripheral blood samples from suspected malaria patients in southeastern Iran, both microscopic and molecular methods were employed, specifically targeting the 18S rRNA gene. Of the 97 samples, 94 were confirmed positive by the microscopic method. In our study, 36 samples tested positive using the microscopic method, while 33 cases were confirmed through molecular analysis (25). In the study conducted by Wang et al., three methods—microscopy, nested PCR, and RT-PCR targeting the 18S rRNA gene—were employed to screen for malaria infections among residents of Myanmar, a malaria-endemic region. The sensitivity results of the two molecular methods were found to be comparable, while the sensitivity of the microscopic method was significantly lower than that of the molecular methods (27). In our study, we also observed a significant difference between the molecular methods and the microscopic method.
In a 2022 study by Kumari et al., the researchers developed a multiplex PCR method utilizing the COX gene to detect P. falciparum and P. vivax. They compared this method with nested 18S rRNA gene and microscopic techniques, reporting higher sensitivity and specificity for their approach (28). It should be noted that our molecular method, based on the 18S rRNA gene, can detect all types of human malaria and can be multiplexed in a single run with high sensitivity and specificity. Another study conducted by Amaral et al. investigated ribosomal and non-ribosomal targets for malaria detection using three methods—nested PCR, real-time PCR, and conventional PCR. They found that, in cases of low parasite infections, the gene targets did not differ significantly. Furthermore, the conventional PCR method utilizing non-ribosomal targets demonstrated the highest sensitivity in diagnosing mixed infections (29).
In other similar research, Demas et al. discovered that specific malaria diagnostic targets, Pvr47 and Pfr364, exhibited greater sensitivity than the 18S rRNA gene. They also observed that multiplex infections could not be detected using the 18S rRNA gene without employing a nested method (8). We have addressed these limitations by developing a multiplex/semi-nested PCR method utilizing the 18S rRNA gene for all human malaria species. In this study, we did not have a positive sample for P. knowlesi, so it was not tested with our custom-designed primers. Our approach, which employs these primers, demonstrates proficient detection of low parasite levels and mixed infections. This is achieved by amplifying gene fragments of varying sizes that can be distinguished through agarose gel electrophoresis. Consequently, this allows for a one-step multiplex application, which is a key strength of this methodology.
5.1. Conclusions
In conclusion, multiplex semi-nested PCR assays have demonstrated satisfactory performance in identifying plasmodia in blood samples. Although this method requires specialized equipment and well-trained personnel and can be time-consuming, multiplex semi-nested PCR assays provide more accurate results than microscopy and RDTs.