1. Background
Klebsiella pneumoniae, a prominent human pathogen within the Enterobacteriaceae family, is recognized for its ability to cause severe hospital-acquired infections worldwide. The pathogen's success is attributed to its diverse virulence factors, biofilm-forming capacity, and genetic variability, which enhance its pathogenicity and resistance to treatment (1-3). Biofilm formation, a critical virulence trait of K. pneumoniae, enables the bacterium to adhere to medical devices and hospital surfaces, leading to persistent infections and treatment challenges (4). The ability to form biofilms is influenced by various factors, including the presence of virulence genes encoding for adhesins (type 1 and type 3 fimbriae), exopolysaccharides, and regulators of biofilm formation (e.g., rmpA and magA) (5, 6).
The diversity of virulence genes among K. pneumoniae strains plays a pivotal role in their pathogenicity and clinical outcomes. Genes involved in iron acquisition systems (entB, ybtS), capsule synthesis (cps loci), and toxin production (e.g., hlyA, allS) contribute to the evasion of host immune responses and disease severity (7, 8). Understanding the prevalence and distribution of these virulence genes among clinical isolates is crucial for assessing the pathogenic potential and epidemiological characteristics of K. pneumoniae strains in hospital settings.
Molecular typing methods such as random amplified polymorphic DNA polymerase chain reaction (RAPD-PCR) offer insights into the genetic diversity and clonal relatedness of K. pneumoniae isolates. This approach facilitates the tracking of outbreak strains, identification of high-risk clones, and understanding of transmission dynamics within healthcare facilities (9). Combining genotyping with the characterization of virulence factors and biofilm formation provides a comprehensive framework for studying the epidemiology and clinical impact of K. pneumoniae infections.
2. Objectives
This study focuses on elucidating the occurrence of virulence genes, RAPD-PCR genotyping, and biofilm formation dynamics among K. pneumoniae isolates collected from hospital units in Amol, northern Iran. By integrating these molecular and phenotypic analyses, we aim to enhance our understanding of the pathogenesis and transmission dynamics of K. pneumoniae in hospital environments, informing strategies for infection control and treatment.
3. Methods
3.1. Klebsiella pneumoniae Isolates and Ethics
Seventy isolates of K. pneumoniae, previously recovered in the bacteriology laboratory of Amol University of Special Modern Technologies, were used. The isolates were obtained from 12 hospital departments: Outpatients, surgery, ICU, infectious, emergency, internal, dialysis, inpatients, pediatric, neurology, CCU, and urology. They were collected from five sample types: Urine culture, sputum culture, abscess drain culture, wound culture, and blood culture. Forty-one isolates were recovered from women, and 29 isolates were recovered from men.
3.2. Template DNA and Capsular Type of the Isolates
Extraction of DNA from K. pneumoniae isolates was carried out using a Gram-negative bacteria DNA purification kit (CinnaClone, Tehran, Iran) following the manufacturer's instructions. The quantification of DNA purity and concentration was conducted using spectrophotometry (Nanodrop 1000, Thermo Scientific) at wavelengths of 260 and 280 nm. Following extraction, the DNA samples were stored at a temperature of -20°C for future use. The capsular type of the isolates was evaluated in a parallel study. According to the method of Zhang et al. (10), PCR detection of seven capsular antigen-related genes, including WzyK1, WzyK2, WzyK3, WzyK5, WzyK20, WzyK54, and WzyK57, identified 52 isolates with the K57 capsule type, three isolates with the K54 capsular type, and 15 isolates with unknown capsular type (unpublished data).
3.3. Biofilm Assay and Hypervirulent Klebsiella pneumoniae Identification
The string test was performed as a screening test for hvKP. Hyperviscosity was confirmed by assessing the production of a mucoviscous cord greater than 5 mm when an inoculation loop was used to span a colony developed on an agar plate (11). After the incubation period, the peg plate used for the biofilm biomass assay was removed from the microtiter plate and thoroughly washed with PBS to eliminate any free-floating cells. The cells were then dried for 30 minutes at 37°C. Each replicate peg underwent staining with 0.5% (w/v) crystal violet for five minutes, after which any excess stain was carefully removed by rinsing the peg plate under running distilled water. The stained biofilms were then decolorized by adding 100 µL of 95% ethanol to each well for one minute. Subsequently, the ethanol solution was transferred to a fresh 96-well microtiter plate for optical density (OD) measurement using a microtiter plate reader.
The biofilm-forming potential of each isolate was evaluated by comparing the absorbance of the crystal violet stain acquired for each biofilm against positive and negative controls. The strains were classified into four distinct categories based on their biofilm formation:
- Absence of biofilm formation (< 25% absorbance relative to the positive control)
- Weak biofilm formation (25% - 50%)
- Moderate biofilm formation (51% - 75%)
- Strong biofilm formation (76% - 100%)
In the biofilm assays, Staphylococcus epidermidis RP62A, a well-known biofilm-forming strain, was used as a positive control, as previously described by Stepanović et al. (12).
3.4. Detection of Virulence Genes
The study analyzed the isolates to detect the presence of seven presumed genes linked to the virulence of K. pneumoniae, including magA (associated with capsular serotype K1 and hypermucoviscosity phenotype), ybtS and entB (related to siderophore production), mrkD (linked to adhesin type 3 fimbriae), rmpA (regulator of mucoid phenotype A), kfu (associated with iron transport and phosphotransferase function), and allS (related to allantoin metabolism). Polymerase chain reaction was carried out using specific primers (Table 1) in a final volume of 25 μL, comprising 12.5 μL of PCR master mix, 1 μL (0.4 μM) of both forward and reverse primers (13, 14), and 2 μL of template DNA. All reagents were procured from CinnaClone Company, Iran.
| Target Genes and Sequence (5' to 3') | Annealing Temperature (°C) | PCR Product Size (bp) | Reference |
|---|---|---|---|
| magA | 53 | 1283 | (13) |
| F: GGTGCTCTTTACATCATTGC | |||
| R: GCAATGGCCATTTGCGTTAG | |||
| ybtS | 52 | 242 | (14) |
| F: GACGGAAACAGCACGGTAAA | |||
| R: GAGCATAATAAGGCGAAAGA | |||
| entB | 60 | 400 | (14) |
| F: GTCAACTGGGCCTTTGAGCCGTC | |||
| R: TATGGGCGTAAACGCCGGTGAT | |||
| mrkD | 57 | 340 | (14) |
| F: AAGCTATCGCTGTACTTCCGGCA | |||
| R: GGCGTTGGCGCTCAGATAGG | |||
| rmpA | 51 | 461 | (14) |
| F: CATAAGAGTATTGGTTGACAG | |||
| R: CTTGCATGAGCCATCTTTCA | |||
| kfu | 53 | 638 | (14) |
| F: GGCCTTTGTCCAGAGCTACG | |||
| R: GGGTCTGGCGCAGAGTATGC | |||
| allS | 55 | 764 | (14) |
| F: CATTACGCACCTTTGTCAGC | |||
| R: GAATGTGTCGGCGATCAGCTT | |||
| RAPD (1254) | 36 | Random | (15) |
| CCGCAGCCAA | |||
| RAPD (1283) | 36 | Random | (16) |
| GCGATCCCCA |
Primers Used in Polymerase Chain Reaction for Identifying Virulence Genes of Klebsiella pneumoniae Isolates
After PCR, the resultant product was assessed through electrophoresis in a 1.5% agarose gel. The identified PCR products were analyzed using a 100 bp DNA ladder (CinnaClone, Iran). Ultimately, distinct patterns of virulence genes were identified based on the presence of these genes.
3.5. DNA Fingerprinting and Phylogenetic Analysis
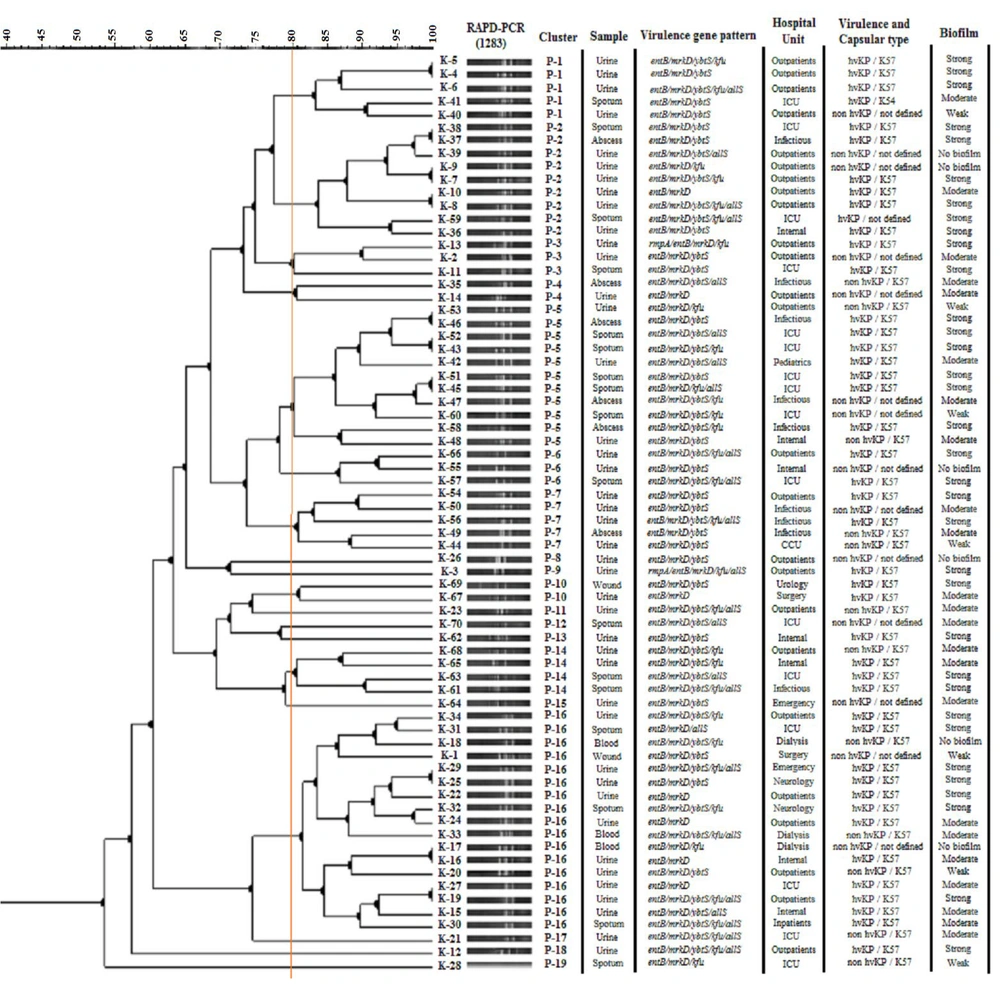
Random amplified polymorphic DNA polymerase chain reaction was carried out using 1254 and 1283 random primers (Table 1), which had been previously characterized (15, 16). The thermocycler (Labnet International, Inc., NJ, USA) followed a prescribed protocol for the RAPD technique: Initial denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 1 minute, annealing at 36°C for 1 minute, and extension at 72°C for 1 minute. The final extension step was conducted at 72°C for 6 minutes. Visualization of the PCR products was performed by gel electrophoresis in a 3% agarose gel. The RAPD reaction images were imported into GelClust software (17). Genetic relatedness was calculated using Pearson's correlation, and the dendrogram was constructed based on the Dice correlation coefficient, along with the unweighted pair group method with arithmetic averages (UPGMA). The final groupings were established by applying a threshold of 80%.
3.6. Statistical Analysis
The outcomes of the research were analyzed using SPSS version 23 software (IBM, Armonk, NY, USA). Various statistical methods, including the Mann-Whitney test, chi-square test, and Kolmogorov-Smirnov test, were employed for the statistical analysis. Significance levels were set at a P-value below 0.05.
4. Results
4.1. Biofilm Formation and Hypervirulent Klebsiella pneumoniae Isolates
Forty-four (62.85%) isolates were identified as hypervirulent K. pneumoniae (hvKP). Among the 70 K. pneumoniae isolates, 64 (91.42%) were biofilm producers. Thirty-three (41.14%), 23 (32.85%), 7 (10%), and 6 (8.57%) isolates exhibited strong, moderate, weak, and no biofilm formation, respectively. Statistically, a significant correlation was detected between hvKP isolates and the capsular type K57 (P < 0.05). The most prevalent phenotype of the isolates was hvKP with the K57 capsular type (Figures 1 and 2). The K54 capsular type, present in three isolates, did not show a significant relationship with other virulence variables. All isolates with a strong biofilm phenotype were hvKP, and this association was statistically significant (P < 0.05).
Dendrogram based on 1254- random amplified polymorphic DNA polymerase chain reaction (RAPD-PCR) fingerprinting of Klebsiella pneumoniae isolates using the unweighted pair group method with arithmetic averages (UPGMA) analysis, in associated with virulence gene pattern, hospital unit, and sample and capsular type
Dendrogram based on 1283- random amplified polymorphic DNA polymerase chain reaction (RAPD-PCR) fingerprinting of Klebsiella pneumoniae isolates using the unweighted pair group method with arithmetic averages (UPGMA) analysis, in associated with virulence gene pattern, hospital unit, and sample and capsular type
4.2. Virulence Gene Detection
The entB and mrkD genes were detected in all 70 isolates. The magA gene was not identified among the isolates. In total, 10 diverse patterns of virulence genes were recognized among the isolates (Table 2). The entB/mrkD/ybtS pattern was the most observed, with a frequency of 21 (30%). This virulence pattern was identified in nine different hospital units. The virulence gene pattern had no significant relationship with other variables, including hvKP status, hospital unit, sample type, gender of the patients, and age group (P > 0.05).
| Number of Genes and Virulence Gene Pattern | Number of Isolates (%) | Sources (n) | Hospital Unit (n) |
|---|---|---|---|
| 2 | |||
| entB/mrkD | 7 (10) | UC (6), SC (1) | Outpatients (4), internal (1), ICU (1), surgery (1) |
| 3 | |||
| entB/mrkD/ybtS | 21 (30) | UC (13), SC (3), AC (3), WC (2) | outpatients (5), internal (4), ICU (3), infectious (4), CCU (1), emergency (1), urology (1), surgery (1), neurology (1) |
| entB/mrkD/kfu | 4 (5.71) | UC (2), SC (1), BC (1) | Outpatients (2), ICU (1), dialysis (1) |
| entB/mrkD/allS | 2 (2.85) | UC (1), SC (1) | Outpatients (1), ICU (1) |
| 4 | |||
| entB/mrkD/ybtS/kfu | 13 (18.57) | UC (5), SC (5), AC (2), BC (1) | Outpatients (4), internal (1), ICU (4), infectious (2), neurology (1), dialysis (1) |
| rmpA/entB/mrkD/kfu | 1 (1.42) | UC (1) | Outpatients (1) |
| entB/mrkD/ybtS/allS | 7 (10) | UC (3), SC (3), AC (1) | Outpatients (1), internal (1), ICU (3), infectious (1), pediatrics (1) |
| entB/mrkD/kfu/allS | 1 (1.42) | SC (1) | ICU (1) |
| 5 | |||
| rmpA/entB/mrkD/kfu/allS | 1 (1.42) | UC (1) | Outpatients (1) |
| entB/mrkD/ybtS/kfu/allS | 13 (18.57) | UC (9), SC (3), BC (1) | Outpatients (6), inpatients (1), ICU (2), infectious (2), emergency (1), dialysis (1) |
| Total | |||
| 10 | 70 | 5 | 12 |
Virulence Gene Patterns of the Klebsiella pneumoniae Isolates Recovered from Hospital Units
4.3. Genotyping Results
The outcomes of the RAPD-PCR are presented in Figures 1 and 2, depicted as dendrograms along with the virulence gene patterns, sample type, hospital unit, and capsular type of the isolates. A total of 25 separate clusters were identified through the examination of 1254 primer RAPD-PCR using GelClust (UPGMA), denoted as R-1 to R-25 (Discriminatory power: 0.9573; SID: 0.04265). A total of 19 separate clusters were identified through the examination of 1283 primer RAPD-PCR using GelClust (UPGMA), denoted as P-1 to P-19 (Discriminatory power: 0.8919; SID: 0.1081).
The 1254-RAPD-PCR showed a statistically significant relationship with sample type (P < 0.05). Some genotypes (R-8, R-9, R-11, R-15, R-17, R-21, R-22, and R-25) belonged to urine culture isolates. Additionally, this genotyping method showed a significant relationship with biofilm production in some genotypes (P < 0.05). No significant relationship was observed between 1254 patterns and the presence of virulence genes or virulence gene patterns (P > 0.05).
The 1283-RAPD-PCR showed a statistically significant relationship with virulence gene patterns and the existence of the rmpA and ybtS genes (P < 0.05). Specifically, the ybtS gene showed a statistically significant relationship with P-1, P-5, P-7, and P-14 (P < 0.05), and the rmpA gene presented a statistically significant relationship with P-3 and P-9 (P < 0.05). In addition, genotypic patterns and clusters obtained from the two primers were significantly related to each other (P < 0.05). These genotypic patterns did not show a significant relationship with other variables.
5. Discussion
The study examined the patterns of virulence genes and biofilm formation capabilities across various capsular and phylogenetic types of K. pneumoniae strains isolated from different hospital units. These findings provide valuable insights into the pathogenicity and epidemiological characteristics of K. pneumoniae, a significant pathogen responsible for severe hospital-acquired infections. The capability of K. pneumoniae to form biofilms is a critical factor in its pathogenicity and antibiotic resistance, as it protects bacterial communities from the host immune response and antimicrobial agents (18). Vuotto et al. showed that extensively drug-resistant (XDR) strains are associated with biofilm production (19).
The current study was conducted in parallel with another study on the antibiotic resistance of this bacterium and focused on the virulence characteristics of the bacteria. Our results showed that a very high proportion of the isolates were found to produce biofilm, and 62.85% of hospital-recovered K. pneumoniae isolates demonstrated strong biofilm formation. In addition, the biofilm formation potential varied significantly among phylogenetic groups, with the majority of strong biofilm producers found in P-2, P-5, and P-16 genotypes of the 1283-RAPD-PCR. This distribution highlights the importance of specific phylogenetic backgrounds in biofilm formation and persistence in clinical settings.
Although the presence of the virulence genes investigated in this study did not demonstrate a significant difference among strains with varying biofilm types, this finding does not necessarily negate the role of these genes in biofilm formation. It is possible that other genetic or environmental factors, such as regulatory pathways or external stimuli, influence biofilm development in K. pneumoniae. Additionally, the lack of significant differences may reflect the complex and multifactorial nature of biofilm formation, where the interplay between virulence factors, host interactions, and microenvironmental conditions collectively determine the biofilm phenotype. Further studies focusing on gene expression levels and functional analyses could provide deeper insights into these relationships (19).
All RAPD genotypes (both 1254 and 1283) demonstrated the ability to produce some form of biofilm, with the exception of cluster R-18 identified in 1254-RAPD-PCR. In a prior study, Seifi et al. reported a correlation between sample type and strong biofilm production (20). However, this relationship was not confirmed in the current study. The discrepancy might be attributed to differences in study design, strain selection, or environmental conditions. Further research focusing on these variables could clarify the factors influencing biofilm formation in K. pneumoniae.
The analysis of the presence of virulence genes revealed noteworthy diversity in the distribution of seven putative virulence genes among K. pneumoniae isolates. All isolates harbored the entB gene related to siderophore production and the mrkD gene related to type 3 fimbriae, indicating the ubiquitous presence of enterobactin synthesis and fimbriae-associated genes, which are essential for iron acquisition and adherence to host cells, respectively (21). Adhesion plays a critical role in establishing infections, particularly in the urinary and respiratory tracts. A study by de Astorza et al. demonstrated that fimbriae-associated genes, such as mrkD, were commonly present in clinical isolates of K. pneumoniae. These genes are essential for bacterial adherence and biofilm formation (22). Additionally, their findings revealed a correlation between the presence of these genes and increased resistance to phagocytosis by host immune cells, thereby enhancing bacterial survival and contributing to its virulence (22). This underscores the importance of targeting adhesion mechanisms in developing strategies to combat K. pneumoniae-associated infections.
In K. pneumoniae, the ybtS gene, a phenolate-type siderophore that is structurally dissimilar from enterobactin and Kfu, facilitates the uptake of ferric iron (14). Notably, in the present study, the ybtS gene was found in a relatively high number of the isolates (77.14%), while the kfu and allS genes, respectively linked to iron uptake and allantoin metabolism, were present in 41.14% and 32.85% of the isolates. Yu et al. previously demonstrated that strains carrying the rmpA gene were associated with hypermucoviscous phenotypes and the clinical syndromes caused by invasive Klebsiella pneumoniae strains (23). In the current study, the rmpA gene was rare, being detected in only 2.85% of the isolates. Notably, both of these isolates were classified as hvKP and exhibited a strong biofilm phenotype. This finding aligns with the established role of rmpA in enhancing capsule production and promoting virulence, further emphasizing its significance in the pathogenicity of hvKP strains.
Dwi Fatmawati et al. concluded that RAPD-PCR possesses significant discriminatory power among K. pneumoniae isolates and can be an effective tool for defining clonal associations and tracking the spread of outbreak strains in hospital units (24). Previous studies have also highlighted the RAPD technique as a more accessible and cost-effective method, providing reliable results in differentiating strains, particularly in laboratories of developing countries with limited access to advanced technologies (25-27).
According to the results, phylogenetic analysis categorized the isolates into distinct groups with varying virulence gene profiles. The P-2 and P-16 clusters of the 1283-RAPD-PCR contained the highest number of isolates with multiple virulence genes, suggesting that these groups might possess enhanced pathogenic potential. Interestingly, the presence of the rmpA gene was limited to a few isolates within the P-3 and P-9 groups, which aligns with the hypermucoviscous phenotype and suggests a potential increase in virulence (Figure 2). The statistical analysis of genotyping results revealed that the genotypic patterns obtained from 1254-RAPD-PCR are suitable and reliable for separating, identifying, and comparing biofilm patterns as well as determining the type of sample, especially urine culture. Additionally, the results showed that 1283-RAPD-PCR genotypic patterns can be used to identify isolates with specific virulence genes (ybtS and rmpA) or specific virulence patterns.
The virulence gene patterns and biofilm formation capabilities were also analyzed concerning the source of the isolates. Isolates from the ICU exhibited a higher prevalence of multiple virulence genes and strong biofilm formation. Specifically, the entB/mrkD/ybtS/kfu/allS gene pattern was predominant in ICU isolates, indicating a potential link between these virulence factors and the severity of infections in critically ill patients. The presence of multiple virulence genes and strong biofilm formation capabilities in K. pneumoniae isolates from critical hospital units underscores the need for stringent infection control measures (28). The high occurrence of entB and mrkD genes in all isolates indicates potential for effective therapeutic interventions. Finally, identifying phylogenetic groups with increased virulence can assist in targeted surveillance and containment efforts.
5.1. Conclusions
This study emphasizes the high prevalence of hvKP strains, particularly those producing strong biofilms and possessing the K57 capsular type, in clinical isolates from hospitals in Amol, northern Iran. The strong association between biofilm production and hvKP underscores the pathogen's enhanced capability for persistence and resistance in hospital settings. Although the entB and mrkD virulence genes were universally present, the diversity of virulence gene patterns and their lack of significant relationships with hvKP status, hospital units, and sample types highlight the complex interplay of factors influencing the pathogenicity of K. pneumoniae.
Random amplified polymorphic DNA polymerase chain reaction fingerprinting revealed considerable genetic diversity among the isolates, identifying unique genotypic clusters that correlated significantly with biofilm formation, sample types, and specific virulence gene patterns. These findings underline the critical role of integrating molecular typing and phenotypic characterization in monitoring K. pneumoniae epidemiology. The study's insights into the genetic and phenotypic traits of hvKP provide valuable information for infection control and therapeutic strategies. Continuous surveillance and targeted interventions are imperative to mitigate the risks associated with hypervirulent and multidrug-resistant K. pneumoniae strains in healthcare environments. Future research should explore the molecular mechanisms driving biofilm formation and virulence in hvKP to develop effective preventive and therapeutic measures.