1. Background
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has emerged as a global pandemic since its first reported case in December 2019 in Wuhan, China (1). This novel respiratory illness presents with symptoms ranging from mild flu-like symptoms to severe pneumonia, leading to critical illness and death in some cases (2). As the virus rapidly spread across the globe, it became evident that co-infections with various pathogens, including bacteria, are common in patients with COVID-19 (3). Among the bacterial co-infections observed in COVID-19 patients, Mycoplasma pneumoniae has been identified as a significant pathogen associated with the disease (4, 5). Mycoplasma pneumoniae is one of the major causative pathogens of community-acquired pneumonia (CAP) and the most frequent pathogen in atypical pneumonia infections (6). Several studies reported that M. pneumoniae is responsible for around 10 - 30% of CAP cases (7-9). In severe cases, M. pneumoniae infection can lead to extrapulmonary manifestations such as myocarditis, nephritis, and hemolytic anemia (10). The CAP caused by atypical pathogens can have symptoms and presentations similar to COVID-19, making it difficult to differentiate between the two based on clinical observations or imaging results (11).
The pathogenesis of M. pneumoniae involves adherence to the host's respiratory tract cells, leading to damage to the respiratory cilia and destruction of the epithelium (12). One of the key adhesins involved in this process is the P1 protein, which, along with P40/P90, forms the transmembrane adhesion complex at the tip of the attachment organelle (13). Understanding the molecular mechanisms of M. pneumoniae virulence is crucial for elucidating its role in co-infections with SARS-CoV-2 and the pathogenesis of COVID-19 (14, 15). Current research indicates that individuals with COVID-19 who are also infected with other respiratory pathogens may experience more severe symptoms and higher mortality rates (16).
Therefore, it is important to conduct studies to identify these co-infections and assess how they impact the overall clinical outcome of COVID-19 patients. In this study, by investigating the co-infection of M. pneumoniae with SARS-CoV-2, we seek to elucidate the clinical implications and risks associated with this bacterial pathogen in COVID-19 patients. Specifically, this study focuses on the molecular characterization of the key virulence genes p1, p40, and p90, which play a critical role in the pathogenesis of M. pneumoniae respiratory infections.
2. Objectives
By advancing our understanding of the interactions between M. pneumoniae and SARS-CoV-2, this study aims to provide insights into the potential implications of bacterial co-infections in COVID-19 patients and the development of targeted therapies for improved patient outcomes.
3. Methods
3.1. Study Design and Sampling
In this study, we conducted a descriptive study to investigate the prevalence of M. pneumoniae and virulence genes in admitted patients with SARS-CoV-2 infection. The study was carried out in three hospitals in Kerman, Iran, from January 2021 to June 2021. The study included a total of 200 patients with COVID-19 (patient group) and 200 healthy individuals who tested negative for SARS-CoV-2 (control group). The age and blood pressure levels of the participants in both groups were studied as dependent variables, with gender, occupation, body weight, and other confounding variables controlled for. The number of women and men selected was equal. None of the participants had a high-risk occupation related to respiratory diseases, and their Body Mass Index (BMI) ranged from 18.5 to 25, which is considered the normal weight range.
Samples were collected from individuals who visited the hospitals over six months in 2021. The age and blood pressure of the patients in both groups were also recorded. The confirmation of positive and negative results in both groups was conducted by the hospital laboratory, followed by repeat real-time PCR testing. The respiratory secretion samples collected from patients included samples from both the upper respiratory tract (pharynx) and lower respiratory tract (lungs). The study included both acute and chronic cases of infection in both the patient and control groups. Clinical symptoms observed in patients ranged from fever, headache, cough, weakness, and lethargy to allergies, diarrhea, and muscle pain. The details and number of collected samples are presented in Table 1 and Appendix 1 in Supplementary File.
| Groups | Number of Samples | Sample Source | |
|---|---|---|---|
| Case group (patients with coronavirus) | |||
| Acute | 100 | Lung (n = 50) | Pharynx (n = 50) |
| Chronic | 100 | Lung (n = 50) | Pharynx (n = 50) |
| Control group (non-coronavirus patients) | |||
| Acute | 100 | Lung (n = 50) | Pharynx (n = 50) |
| Chronic | 100 | Lung (n = 50 | Pharynx (n = 50) |
| Total | 400 | ||
3.2. Polymerase Chain Reaction Analysis
3.2.1. DNA Extraction
The DNA extraction process involved isolating the DNA from each sample using the CinnaPure-DNA purification kit (CinnaGen, Iran) according to the manufacturer's instructions and was used as the template for PCR amplification.
3.2.2. Specific Primer Design and Gene Targets
The specific primers to amplify target genes for the identification of the Mycoplasma genus, as well as the p1, p40, and p90 virulence genes of M. pneumoniae, were analyzed and designed according to gene information in the GenBank database using the MEGA5 software (Table 2).
| Primer | Sequence (5'-3') | Length (bp) |
|---|---|---|
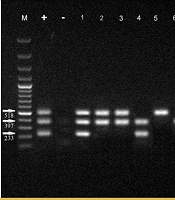
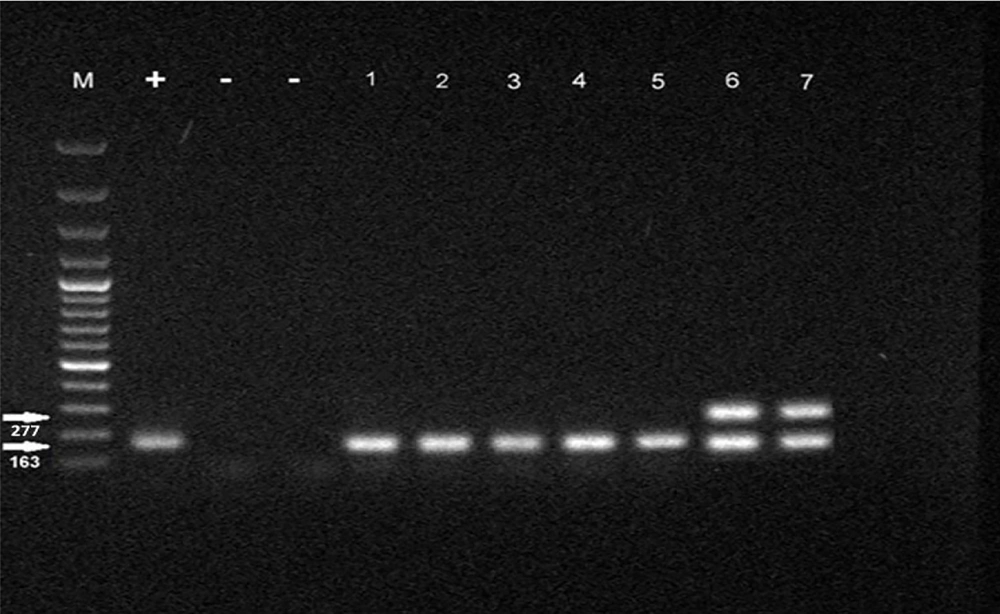
| MH1, MH2 | F: TGAAAGGCGCTGTAAGGCGC; R: GTCTGCAATCATTTCCTATTCCAAA | 277 |
| MG37 | F: AGTTGATGAAACCTTAACCCCTTGG; R: CCGTTGAGGGGTTTTCCATTTTTGC | 163 |
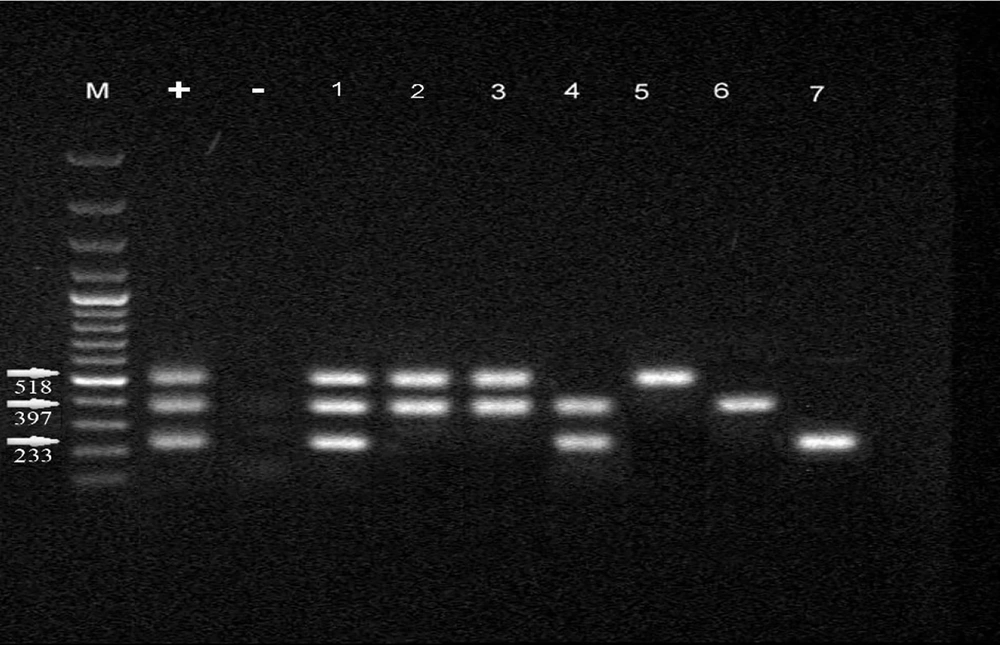
| P1 | F: TTATTGAAATACGATACAGCTCATGGA; R: CGTTGGCAATAGGAGCTAAACAG | 233 |
| P40 | F: TAAGAGTTTGATCCTGGCTCAG; R: GGACTACCAGGGTATCTAATCC | 397 |
| P90 | F: TGCAATCTGCTCGTGAAGTATTAC; R: CGAAACGACGTCCATAAGCAACT | 518 |
a Target gene: 16s rRNA
3.2.3. PCR Amplification
The PCR method was conducted on total volumes of 25 μL using 12 μL of mixed master mix (Sinaclon, Iran), 2 μL of DNA, 1 μL of each primer (20 pmol/μL), and 9 μL of double distilled water (DDW). The amplification conditions for the Mycoplasma genus and species using specific primers were processed by initial heating at 94°C for 7 minutes, 30 cycles of denaturation at 94°C for 30 seconds, annealing at 62°C for 30 seconds, and extension at 72°C for 30 seconds, followed by a 5-minute extension at 72°C. In addition, p1, p40, and p90 virulence genes detection was carried out by initial heating at 35°C for 5 minutes, 35 cycles of denaturation at 95°C for 45 seconds, annealing at 55°C for 40 seconds, and extension at 72°C for 40 seconds, followed by a 5-minute extension at 72°C. PCR products were analyzed by electrophoresis (50 V, 2 h) in 1% agarose gels stained with 0.5 μL/mL of ethidium bromide. After that, PCR products were visualized and photographed using a GEL Imaging System (Bio-Rad Laboratories, CA, USA).
3.3. Statistical Analysis
Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS 10.0 for Windows). The chi-square test was used with a significance level (α) of 0.05. The confidence interval was considered to be 95%.
4. Results
4.1. PCR Amplification
In total, 400 samples of respiratory secretions were used in the current study, of which 200 patients were positive for genome targets of SARS-CoV-2 by real-time PCR, and 200 patients were negative. All samples were subjected to PCR amplification for the Mycoplasma genus and virulence genes of M. pneumoniae in both case and control groups. A total of 44 (11%) and 35 (8.75%) samples were positive for the Mycoplasma genus and M. pneumoniae, respectively, according to the PCR analysis in both groups. The PCR analysis results are shown in Figures 1 and 2.
4.2. Frequency of Mycoplasma Genus in Chronic and Acute Samples of Lung and Pharynx
Based on the data analysis, the study found that there was no significant difference in the proportion of Mycoplasma genus infection rates between the control groups with acute and chronic lung infection and those infected with coronavirus (P = 0.734). In the control group, 6% of individuals with acute lung damage and 26% of individuals with chronic lung damage were infected with the Mycoplasma genus. Among individuals infected with coronavirus, 12% of those with acute lung damage and 22% with chronic lung damage were infected with the Mycoplasma genus. Additionally, the proportion of individuals infected with the Mycoplasma genus was relatively consistent in both the lung and pharyngeal samples across the different groups. Comparing the number of Mycoplasma in lung samples from control subjects with acute and chronic lung injury revealed a significant difference between the two groups. Specifically, samples from control subjects without exposure to COVID-19 showed a higher percentage of Mycoplasma species in samples with chronic lung damage compared to those with acute lung damage (P = 0.012). The results of Mycoplasma genus isolation in the control and case groups are presented in Table 3 and Appendix 1 in Supplementary File.
| Infection | Case Group | Control Group |
|---|---|---|
| Acute lung | 6 (12) | 3 (6) |
| Chronic lung | 11 (22) | 13 (26) |
| Acute pharyngeal | 3 (6) | 3 (6) |
| Chronic pharyngeal | 3 (6) | 2 (4) |
a Values are expressed as No. (%).
4.3. Frequency of Mycoplasma pneumoniae in Chronic and Acute Samples of Lung and Pharynx
Based on the prevalence of the p1, p40, and p90 virulence genes among the control samples, it was found that a higher proportion of individuals with chronic lung damage were infected with M. pneumoniae compared to those with acute lung damage. However, there was no significant difference in the prevalence of infection between the control and coronavirus groups. Further analysis comparing the presence of M. pneumoniae in lung samples from control subjects with acute and chronic injury revealed a significant difference between the two groups.
In individuals without exposure to COVID-19, samples showing chronic lung damage exhibited a higher percentage of M. pneumoniae compared to samples with acute lung damage (P = 0.008). Notably, M. pneumoniae was not detected in pharyngeal samples from control subjects with acute and chronic disease. Additional details regarding these findings can be found in Table 4. The difference in the p virulence gene in both control and case groups showed that there is no significant difference in the distribution of these genes between the two groups (P = 0.708) (Figure 3).
| Infection | Case Group | Control Group |
|---|---|---|
| Acute lung | 6 (12) | 2 (4) |
| Chronic lung | 10 (20) | 12 (24) |
| Acute pharyngeal | 3 (6) | 0 (0) |
| Chronic pharyngeal | 2 (4) | 0 (0) |
a Values are expressed as No. (%).
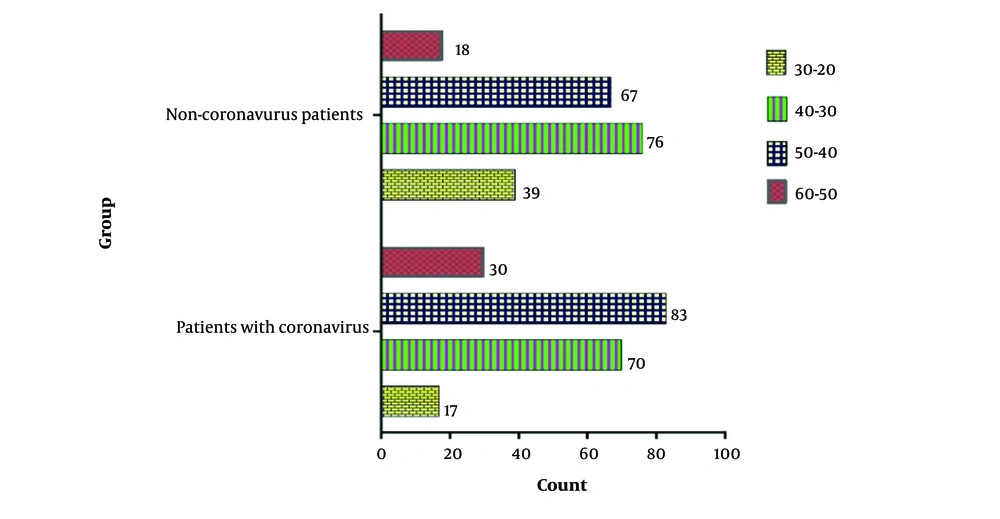
4.4. Age and Blood Pressure Effect
The chi-square test analysis of the blood pressure of individuals in both the control and patient groups revealed no significant difference in the prevalence of high blood pressure (P = 0.695). Additionally, the age distribution of people in both control and patient groups was analyzed by the chi-square test. The result of the test showed that there is a significant difference in terms of age distribution between the two groups (P = 0.004). Based on the results, it was observed that in the patient group, the age group of 40-50 years had a higher percentage of Mycoplasma isolation compared to the control (Figure 4).
5. Discussion
The emergence of the COVID-19 pandemic has brought significant challenges to the healthcare system worldwide (17). Understanding the impact of bacterial co-infections in COVID-19 patients is crucial for determining optimal treatment strategies and improving patient outcomes (18). To the best of our knowledge, there are few molecularly documented cases of co-infection with both SARS-CoV-2 and M. pneumoniae in the medical literature, and co-infection of Mycoplasma with SARS-CoV usually has been found in serological tests (4, 19). In this study, we investigated the co-infection of Mycoplasma and virulence p genes in COVID-19 and non-COVID-19 patients. The results of our study revealed that approximately 11% of respiratory secretion samples from both COVID-19 patients and non-coronavirus individuals were positive for the Mycoplasma genus, with a slightly higher prevalence of M. pneumoniae in the case group. This finding highlights the potential role of M. pneumoniae as a common co-infecting pathogen in individuals with COVID-19, which could exacerbate respiratory symptoms and lead to more severe clinical outcomes (20). High rates of COVID-19-bacterial co-infections identified among COVID-19 patients have also been reported by other researchers (7, 21-23).
Furthermore, this study found that individuals with chronic lung damage, both in the COVID-19 patient group and the control group, had a higher prevalence of M. pneumoniae infection compared to those with acute lung damage. This suggests that chronic respiratory conditions may predispose individuals to bacterial co-infections, including M. pneumoniae, which could further complicate the course of COVID-19 (24, 25). Moeller et al. suggested that children with bronchopulmonary dysplasia (BPD) and other respiratory conditions were at higher risk of requiring ventilatory support, such as supplemental oxygen or invasive ventilation, if infected with the virus (26). The presence of M. pneumoniae in chronic lung damage samples highlights the need for targeted monitoring and treatment of bacterial pathogens in individuals with underlying conditions. COVID-19 can cause inflammation and damage to the lungs, making it harder for individuals with pre-existing lung conditions to breathe properly. This can lead to a higher risk of developing respiratory failure, pneumonia, and acute respiratory distress syndrome (ARDS) (27).
The analysis of the virulence genes p1, p40, and p90 of M. pneumoniae in our study did not reveal a significant difference in the distribution of these genes between the COVID-19 patient group and the control group. However, the higher prevalence of M. pneumoniae in chronic lung damage samples suggests that the expression of these virulence genes may contribute to the pathogenesis of the disease in individuals with pre-existing respiratory conditions. These findings are consistent with previous studies by Shi et al. (28) and Vizarraga et al. (13). Further studies focusing on the molecular mechanisms of M. pneumoniae virulence and its interaction with SARS-CoV-2 are warranted to elucidate the role of this bacterium in COVID-19 pathogenesis.
The age distribution analysis in the study revealed a higher percentage of Mycoplasma isolation in the age group of 40 - 50 years in the patient group compared to the control group. This finding highlights the potential susceptibility of middle-aged individuals to M. pneumoniae infections, particularly in the context of COVID-19. However, Li et al. found that the positive detection rate of M. pneumoniae was highest among children aged 5 to 7 years old in the outbreak of COVID-19 (22). Age-related factors, such as immune function and comorbidities, may influence the susceptibility to bacterial co-infections in COVID-19 patients and should be considered in clinical management strategies (29, 30).
5.1. Conclusions
This study provides important insights into the prevalence of M. pneumoniae and its virulence genes in COVID-19 patients and highlights the potential clinical implications of bacterial co-infections in individuals with COVID-19. Targeted monitoring and treatment of bacterial pathogens, particularly in individuals with chronic respiratory conditions, may help improve patient outcomes and reduce the burden of co-infections in COVID-19. Further research on the molecular mechanisms of M. pneumoniae virulence and its interactions with SARS-CoV-2 is necessary to advance our understanding of bacterial co-infections in COVID-19 and develop targeted therapeutic interventions for improved patient care. Changes in treatment and care protocols, history of patients' respiratory diseases, and change of seasons were some of the limitations of this research.