1. Background
Staphylococcus aureus is an opportunistic pathogen responsible for a considerable mortality rate. Staphylococcus aureus is associated with the asymptomatic colonization of human skin and mucosal surfaces (1). However, this pathogen is an important cause of wound infections and can lead to bone inflammation, endocarditis, and bacteremia, resulting in secondary infections in major organs. Staphylococcal infections often occur when skin and mucosal barriers are compromised, following defects in foreign bodies and the host's immune defense systems (2). Staphylococcus aureus has recently become more prominent due to the advent of methicillin-resistant S. aureus (MRSA) (3). For instance, its occurrence in common places often requires more sophisticated treatment options. Initially, MRSA was limited to the healthcare sector; hence, it was called healthcare-associated (HA)-MRSA (4). In 1997, the first clinical isolate of S. aureus exhibiting moderate resistance to vancomycin was documented in Japan. Subsequent research has identified the presence of vancomycin-intermediate S. aureus (VISA) strains (with a minimal inhibitory concentration of 8 µg/mL) in some countries, including the United States (5).
Polysaccharide capsules are involved in various disease-related functions and processes, including drying prevention, adhesion, and resistance to host immune defenses. Preventing the activity of neutrophils, chemotaxis, and phagocytosis is one of the most important roles of polysaccharide capsules in S. aureus (6). The effective binding of bacteria to host cells and tissues is essential in the initial establishment of organisms in the host body. Encapsulated S. aureus is protected against host-dependent invasion and spreads rapidly in the host body. The capsular polysaccharides (CP) of S. aureus are classified into 11 types, of which only two, Types 5 and 8 (encoded by cap5 and cap8 genes, respectively), are present in 80 - 90% of clinically important pathogenic strains of MRSA (6). Capsules are suitable vaccine targets against other encapsulated pathogens (7). More than 80% of clinical isolates produce toxins with cap5 or cap8. Recent studies have shown that cap5 plays an essential role in S. aureus pathogenesis, likely allowing it to resist the host immune system and phagocytic absorption and killing (8). The rest of the strains have surface polysaccharide antigens 336, which are known as non-typeable S. aureus (9).
2. Objectives
This study examined clinical isolates obtained from hospitals affiliated with Tehran University of Medical Sciences to determine the frequency of capsule-encoding genes in MRSA isolates.
3. Methods
3.1. Bacterial Isolates
This study was conducted on clinical samples sent to the laboratories of three teaching hospitals affiliated with Tehran University of Medical Sciences, including Shariati Hospital, Imam Khomeini Hospital Complex, and Children's Medical Center Hospital. During the period from 2022 to 2023, clinical samples were acquired from various sources such as blood, cerebrospinal fluid (CSF), wound, trachea, urine, abscess, pleura, and synovial fluid. Culture was performed on blood and Chapman agar media (Merck, Germany). In addition, biochemical tests were performed to identify S. aureus isolates, including Gram-staining, catalase, coagulase, mannitol fermentation, urease, and DNase (Merck, Germany). To identify MRSA isolates, antibiotic resistance to cefoxitin and the presence of the mecA gene were investigated. Furthermore, the minimum inhibitory concentration (MIC) of vancomycin was measured using a vancomycin E-test strip, with ≥ 16 µg/mL considered resistant.
3.2. Antimicrobial Susceptibility Test
Antibiotic sensitivity testing was done by the Kirby-Bauer disk diffusion method following the Clinical and Laboratory Standards Institute (CLSI) 2021 standard protocol (10) using a variety of antibiotic discs, including trimethoprim-sulfamethoxazole (SXT, 23 µg), amoxicillin (An, 25 µg), cefazolin (CZ, 30 µg), ciprofloxacin (CP, 5 µg), ceftriaxone (CRO, 30 µg), imipenem (IPM, 30 µg), erythromycin (E, 15 µg), cefotaxime (CTX, 30 µg), clindamycin (CC, 2 µg), cefoxitin (FOX, 30 µg), vancomycin (V, 30 µg), cloxacillin (CX, 1 µg), and linezolid (LZD, 30 µg). However, cefoxitin was used for the phenotypic detection of MRSA. A standardized bacterial inoculum (usually 0.5 McFarland) was inoculated onto the surface of a Mueller-Hinton (MH) agar plate (Merck, Germany) by lawn culture. The antibiotic discs (Mast, UK) were prepared and placed on the plate with appropriate distances under aseptic conditions. After 24 hours of incubation, the inhibition zone diameter around each antibiotic disc was measured.
3.3. Molecular Identification of mecA, vanA, cap5, and cap8 Genes in MRSA Isolates
3.3.1. DNA Extraction
The boiling method was used for DNA extraction. DNA was extracted after an overnight (24 h) pure culture of the sample on brain heart infusion (BHI) agar (Merck, Germany). Initially, some pure colonies were removed from the BHI agar and suspended in 400 µL of distilled H2O, then inoculated into a pre-sterilized microtube, shaken for a few seconds, transferred to a thermo-block BIONER (ThermoCell HB-202/CHB-202), and incubated at 100°C for 15 minutes. Immediately after boiling, they were cooled in a freezer at -20°C for 2 minutes, and the microtubes were centrifuged at 13000 XG for 10 minutes. After centrifugation, the DNA supernatant was transferred to another sterilized microtube, and the DNA concentration was measured using a thermo-nano drop device (DeNovix DS-11 FX).
3.3.2. Polymerase Chain Reaction
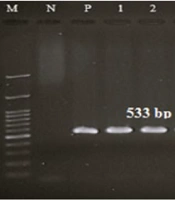
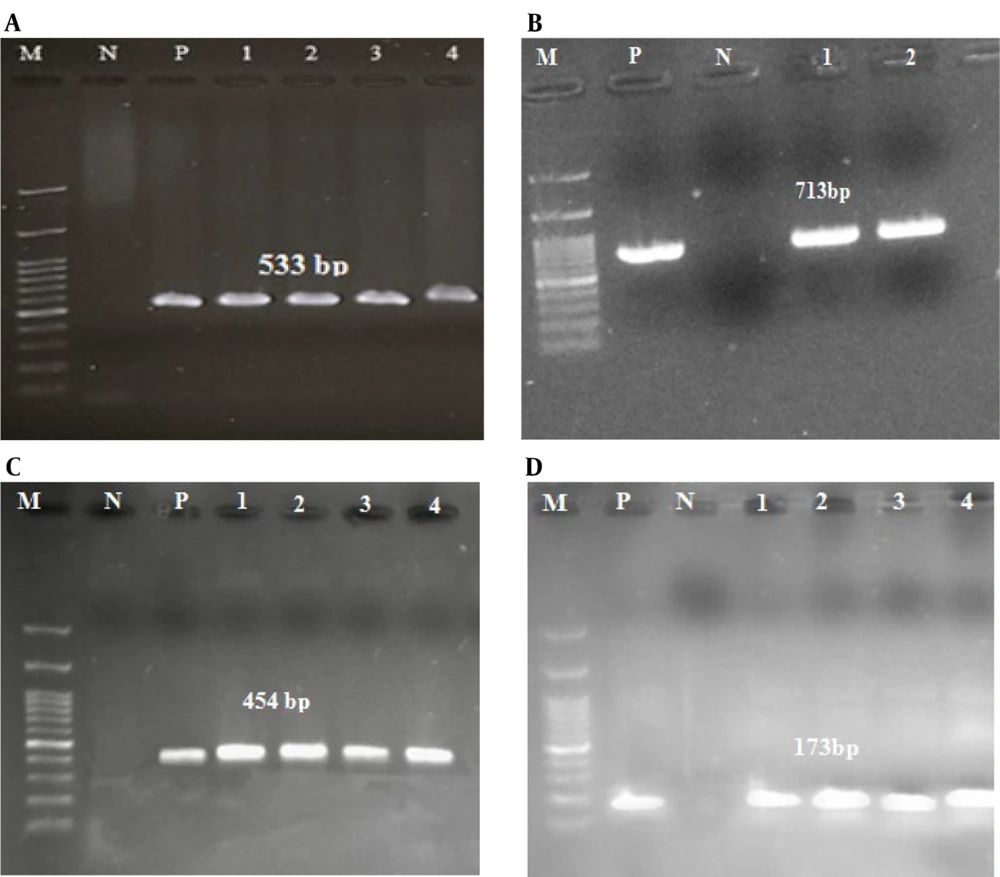
Polymerase chain reaction (PCR) was employed to ascertain the frequency of specific genes in the samples under investigation. To perform the PCR test, the following materials were required: DNA (100 ng/µL, 1.5 µL), distilled water (7.5 µL), master mix ready to use 1X (10 µL), and primers (10 pmol, 1 µL), with a total volume of 20 µL (Table 1). These materials were combined in a PeQlab peQSTAR thermocycler set at appropriate temperature conditions. The pre-denaturation temperature was 94°C to 95°C for 5 minutes, and the denaturation temperature was 95°C for 30 to 40 seconds (based on the gene). The annealing temperature for each gene was set as follows: For mecA at 58°C for 40 seconds, for vanA at 55°C for 30 seconds, for cap8 at 56°C for 30 seconds, and for cap5 at 51°C for 30 seconds. Extension was done at 72°C for 1 minute, with a post-extension step done at 72°C for 5 minutes. Positive control strains for mecA were S. aureus ATCC 25923, for vanA was E. faecium ATCC 51559, and for cap5 & cap8, DNA that contains the gene of interest, which was validated in an internal study, was used.
Specifications of Primers Used for the Studied Genes
3.4. Statistical Analysis
Data were analyzed using SPSS software version 21.
4. Results
4.1. Sample Preparation and Collection
In this study, 73 isolates were confirmed as MRSA. The analysis of hospital departments outlined the ward with the highest prevalence of MRSA clinical isolates. Several hospital departments were investigated. The intensive care unit (ICU) Department had the highest proportion of MRSA clinical isolates (n = 25, 34.24%), while the Ear, Nose, Throat (ENT) Department had the lowest proportion of MRSA clinical isolates (n = 1, 1.36%). The other departments included Burn (n = 16, 22%), Surgery (n = 8, 10.95%), Orthopedic (n = 7, 9.58%), Neurosurgery (n = 5, 6.84%), Oncology (n = 3, 4.10%), Pediatric (n = 3, 4.10%), neonatal ICU (NICU) (n = 3, 4.10%), and Outpatient (n = 2, 2.73%). In this research, all samples were evaluated, and it was found that the collected MRSA isolates from males (n = 42, 57.54%) were more than those collected from females (n = 31, 42.46%). However, no notable correlation was identified between gender and the prevalence of MRSA infection (P = 0.1). As observed in Table 2, the abundance of MRSA isolates was highest in wound samples (n = 21, 28.77%) and lowest in synovial fluid samples (n = 1, 1.36%).
| Samples | No. (%) |
|---|---|
| Wound discharge | 21 (28.77) |
| Blood | 20 (27.40) |
| Trachea | 10 (13.70) |
| Urine | 9 (12.33) |
| Abscess | 6 (8.22) |
| CSF | 4 (5.48) |
| Pleura infusion | 2 (2.74) |
| Synovial fluid | 1 (1.36) |
Frequency Distribution of Methicillin-Resistant Staphylococcus aureus Isolates in Different Clinical Samples
4.2. Antimicrobial Susceptibility Testing
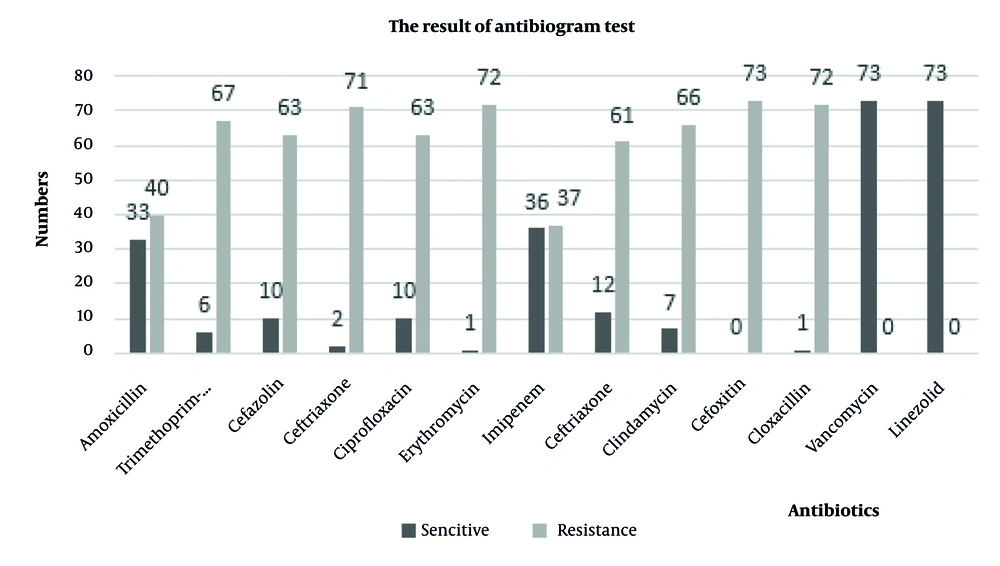
Kirby-Bauer antibiotic susceptibility testing was conducted using antibiotics listed in the CLSI 2021 standards. The results demonstrated that 100% of the strains were susceptible to vancomycin and linezolid, and 100% were resistant to cefoxitin (Figure 1). In this study, the molecular characteristics of MRSA isolates and the presence of vanA, cap5, and cap8 genes were investigated. All the strains confirmed to be cefoxitin-resistant contained mecA. Among the confirmed MRSA isolates, five (6.84%) isolates contained vancomycin resistance genes and were identified as VRSA. The frequency of cap5 and cap8 genes in MRSA isolates was 32 (43.83%) and 27 (36.99%), respectively. No cap5 and cap8 genes were observed in 14 (19.18%) isolates. Out of the five VRSA isolates, two (40%) isolates contained both cap5 and cap8 genes simultaneously, one (20%) isolate contained cap5, one (20%) isolate contained cap8, and one (20%) isolate contained none of the capsular genes. Among the clinical isolates studied, two S. aureus isolates from blood and CSF samples contained all of the mecA, vanA, cap5, and cap8 genes simultaneously. The gel electrophoresis image of the studied genes is shown in Figure 2.
5. Discussion
Methicillin-resistant S. aureus causes several difficult-to-treat infections in humans and resulted in over 100,000 deaths worldwide due to antimicrobial resistance in 2019. In the United States, community-associated (CA)-MRSA leads to an annual economic burden ranging from $478 million to $2.2 billion for third-party payers, and from $1.4 billion to $13.8 billion for society, depending on the definitions and incidences of CA-MRSA (14). The abundance of MRSA isolates was highest in wound samples (n = 21, 28.77%) and lowest in synovial fluid samples (n = 1, 1.36%). The prevalence of MRSA strains was highest in the ICU department (n = 25, 34.24%) and lowest in the ENT department (n = 1, 1.36%).
In the present study, no significant correlation was found between gender and MRSA infections. Similarly, Mohraz et al. identified no significant relationship between gender and MRSA infections (15). In South Korea, the prevalence of MRSA among pediatric patients was notably elevated, ranging from 65.2% to 76.1% between 2002 and 2016 (16). In another study by Tabaei et al. in Mashhad, out of 7,335 bacteria isolated from patients hospitalized in Imam Reza (AS) hospital in Mashhad, the prevalence of MRSA was 41.7% (n = 382). Most MRSA isolates were obtained from blood and wound culture samples (17). Additionally, in studies conducted in Saudi Arabia and Kuwait, this prevalence has been reported to be 11% and 12%, respectively (18, 19).
The capacity of MRSA to induce multiple infections is attributed to the expression of a plethora of pathogenic factors, including adhesins, cytotoxins, superantigens, exoenzymes, and CP, all of which contribute to the pathogenesis of staphylococcal infections (4). The accessory gene regulator (agr) is a key regulatory system that controls the expression of virulence factors in S. aureus. There are two main categories of virulence factors associated with the agr response: Cell wall-associated factors and the regulation of exoprotein secretion. The first category of virulence factors aids in host attachment, helping the bacteria evade the immune system. The second category enhances the expression of polysaccharide capsules and various extracellular toxins, such as hemolysins, enterotoxins, and extracellular proteases (20). The capsule is an essential pathogenic factor in this bacterium, protecting it from phagocytosis and neutrophil activity by clearing the cells. Capsular polysaccharides are polymer structures that encase the cell wall of MRSA. Studies indicate that between 76% and 90% of clinical MRSA isolates produce CP. These CP enhance the virulence of S. aureus by hindering complement and antibody-driven opsonization, as well as by inhibiting phagocytosis (21).
In the present study, 32 (43.83%) isolates contained cap5, and 27 (32.53%) isolates contained cap8; additionally, 14 (19.18%) isolates had no cap5 and cap8 genes, and nine (12.32%) isolates contained both cap5 and cap8 genes. The isolates were obtained from blood, wound, and CSF samples. The distribution of polysaccharide capsules has been analyzed in other countries worldwide, and the distribution percentage varies significantly between studies. In Kuwait in 2020, the prevalence of the cap8 gene was 3.8% in the examined samples (22). Additionally, Liu et al. reported that 81 (56.64%) strains were identified as cap5 genotype, 36 (25.17%) strains were classified as cap8 genotype, and the remaining 26 (18.18%) strains were untypeable (23). Verdier et al. in France indicated that 42% (n = 195) of S. aureus isolates were identified as cap5 genotype, 45% as cap8 genotype, and 13% as other types (8). Given that the strains expressing type 5 and 8 polysaccharide capsules account for approximately 80% of clinical isolates, the role of these capsules in pathogenicity remains a topic of contention. However, recent research has indicated that type 5 capsules may serve as a target for antibodies that protect against experimental S. aureus infections. Consequently, given their prevalence among clinical isolates, they may be considered a viable option for their influential role (22).
The current prevalence of MRSA strains represents a significant public health concern. In response to the pervasive emergence of MRSA, the therapeutic approach to MRSA infections has been modified to include vancomycin. Despite its efficacy in combating MRSA infections, there is a concerning rise in the prevalence of S. aureus strains exhibiting reduced susceptibility to vancomycin and other glycoproteins. The results of the antimicrobial tests conducted in the present study demonstrated the presence of multidrug resistance in the isolates under investigation. All MRSA isolates showed resistance to cefoxitin and sensitivity to vancomycin and linezolid. Our present study identified five (6.84%) MRSA isolates that exhibited vancomycin resistance. In a study by Rezazadeh et al., all MRSA strains were resistant to penicillin and sensitive to mupirocin (24). Askari et al. showed the highest resistance to penicillin (96.6%) and erythromycin (45%) among MRSA strains isolated from clinical samples (25). A study conducted in Nigeria demonstrated that 272 (76.6%) out of 355 S. aureus isolates exhibited susceptibility to vancomycin (26). In 2016, 1,360 samples were examined in Ethiopia, of which 194 were identified as S. aureus. Of these, 34 (17.5%) were determined to be MRSA, and 10 cases (5.1%) exhibited resistance to vancomycin (27).
The data indicate that the main reason for the increase in the prevalence of MRSA strains and the decrease in the prevalence of sensitive strains in hospitals may be attributed to various unnecessary antibiotic prescriptions to patients in hospitals or the community. Consequently, several specific genes in MRSA strains might have adapted to hospital environments, enabling them to persist more effectively than other strains grown in hospitals. A three-year study conducted in Iran on the prevalence of vancomycin-resistant S. aureus isolates between 2014 and 2017 demonstrated that four (0.2%) out of 1,798 samples were VRSA, while two isolates (0.1%) exhibited VISA characteristics (28). In a study by Havaei et al. in Isfahan, the majority of clinical isolates (73%) expressed capsular polysaccharide types 5 (24%) and 8 (49%), while 27% were untypeable (13).
The distribution of MRSA and VRSA strains varies across different geographical areas, and the pattern of sensitivity of these strains to antimicrobial agents is changing. The frequency of MRSA strains from hospital infections and clinical cases, the prevalence of multiple antibiotic resistance in these strains, and the emergence of VISA and VRSA strains in hospitals pose a severe threat regarding therapeutic risks and non-response to microbial treatments (29). At least 13 types of SCCmec elements have been identified, and various clones of MRSA have been observed among epidemic MRSA isolates. Successful lineages of epidemic MRSA clones may have an adaptive advantage due to their antibiotic resistance and virulence (6). The current prevalence of MRSA strains represents a significant public health concern. In response to the pervasive emergence of MRSA, the therapeutic approach to MRSA infections has been modified to include vancomycin. Since the 1980s, vancomycin has been considered the preferred agent for the treatment of severe MRSA infections in numerous healthcare settings. Despite its efficacy in combating MRSA infections, there is a concerning rise in the prevalence of S. aureus strains exhibiting reduced susceptibility to vancomycin and other glycopeptides (30).
5.1. Conclusions
This study, along with numerous previous investigations, demonstrates that the prevalence of cap5 in MRSA clinical isolates is exceedingly high, and the prevalence of cap8 in MRSA clinical isolates was almost similar to previous studies. Identification of capsular genotypes of MRSA clinical isolates is crucial, as CP are integral components of potential vaccine formulations against S. aureus. One strategy for preventing staphylococcal infections is the development of vaccines, in which CP play an important role. Specific antibodies targeting CP5 and CP8 have been shown to provide protection against S. aureus infections. Furthermore, they may serve as promising strategies in the future to combat MRSA and drug-resistant VRSA strains. Results of antimicrobial susceptibility testing in numerous studies indicate that linezolid may be effective against VRSA strains. It would be prudent to implement continuous and nationwide monitoring programs to ascertain vancomycin sensitivity patterns in our country.