1. Background
Klebsiella pneumoniae is a prevalent Gram-negative bacterium encountered in clinical practice, responsible for various infections, including pneumonia, urinary tract infections, sepsis, wound infections, and meningitis (1). The alarming rise in carbapenem-resistant Klebsiella pneumoniae (CRKP) has been attributed to the widespread use of carbapenems in clinical settings (2). Data from the China Antimicrobial Surveillance Network (CHINET) (3) indicate a significant increase in resistance rates of K. pneumoniae to imipenem and meropenem, rising from 3.0% and 2.9% in 2005 to 23.0% and 24.4% in 2021, respectively. The CRKP infections lead to prolonged disease courses, increased treatment costs, and higher mortality rates, severely impacting patient prognosis (4). The CRKP has become a global concern and has been designated as a priority urgent bacterial species by the World Health Organization (5).
The emergence of blaKPC-2-producing K. pneumoniae in China was first documented in 2007 in Zhejiang province (6). Since then, the blaKPC-2 gene has progressively disseminated, becoming the predominant carbapenemase gene not only in Zhejiang (7) but also across the entire country (8). Research indicates slight variations in CRKP prevalence among different regions and hospital levels (9). Therefore, a detailed epidemiological study of CRKP in Lishui, Zhejiang province, over the past decade is essential to understanding its transmission dynamics. A deeper understanding and robust monitoring of these isolates are crucial for limiting the spread of antimicrobial resistance and ensuring the sustained efficacy of new antibiotics.
2. Objectives
This study aimed to investigate the prevalence and risk factors of CRKP in Lishui city from 2015 to 2024. Additionally, we analyzed the key mechanisms of carbapenem resistance to inform hospital infection control strategies and guide clinical antimicrobial therapy for CRKP.
3. Methods
3.1. Bacterial Identification and Antimicrobial Susceptibility Testing
Klebsiella pneumoniae strains were isolated from hospitalized patients at a Grade III Class A general hospital in Lishui city, China, between January 2015 and December 2024. Repeat strains from the same patient were excluded. Identification of isolates was performed using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonik GmbH, Bremen, Germany). In brief, single colonies were evenly spread on the MALDI-TOF MS target plate. A 1 µL matrix solution was then spotted onto the sample. The sample and matrix solution were allowed to dry naturally to form crystals. The target plate was subsequently placed into the instrument, which automatically analyzed the mass spectrum of the sample and compared it with known bacterial species in the database. A high-confidence identification result (score ≥ 2.000, green) indicated that the identification was reliable.
Antimicrobial susceptibility testing of K. pneumoniae isolates against commonly used clinical antibiotics was conducted using the broth microdilution method with the VITEK2 Compact automated microbial analyzer (Biomerieux, France). The interpretation of susceptibility results followed the annual M100-S34 guidelines, available on the Clinical and Laboratory Standards Institute CLSI. ATCC 700603 was used as the quality control strain.
3.2. Carbapenem-Resistant <i>Klebsiella pneumoniae</i> Criteria
According to the Clinical and Laboratory Standards Institute (2024), CRKP was defined as any isolate with a minimum inhibitory concentration (MIC) of imipenem ≥ 4 μg/mL, meropenem ≥ 4 μg/mL, or ertapenem ≥ 2 μg/mL (10).
3.3. Whole Genome Sequencing
Test bacteria were cultured on Mueller-Hinton (MH) plates, and genomic DNA was extracted using a genome extraction kit before being sent to a professional institution for gene sequencing analysis. Data assembly was conducted using SPAdes software, while Prokka 1.5 was employed for gene annotation. Antibiotic resistance genes were identified through the ResFinder 4.1 database. Multilocus sequence typing (MLST) was performed using MLST 2.0 to determine strain sequence types.
3.4. Fourier-Transform Infrared Spectroscopy
Fourier-transform infrared spectroscopy (FTIR) is a phenotypic detection method that generates specific fingerprints by analyzing the absorption of infrared light by various molecular components, including lipids, carbohydrates, and lipopolysaccharides. Recently, FTIR has been utilized for typing K. pneumoniae (11). The specific steps involve scraping a ring of fresh colonies from an overnight culture on MH plates using a 10 μL inoculating loop into a 1.5 mL EP tube containing 100 μL of 70% ethanol, followed by thorough shaking to create a suspension. Next, 100 μL of deionized water was added to each ring and mixed well. A 15 μL suspension was then transferred to a 96-well silica sample plate (Bruker Daltonik, Germany) and dried at 37°C until film formation. Four parallel tests were conducted for each sample. On-machine detection and analysis were performed using the IR Biotyper according to the instrument instructions.
3.5. Statistical Analysis
The statistical data were presented as the number of cases, odds ratio (OR), and percentage. Binary logistic regression was utilized to evaluate the OR and 95% confidence interval (CI) for the univariate analysis of CRKP-related risk factors, with P < 0.05 considered statistically significant.
4. Results
4.1. General Situation of Hospital Monitoring Data
Klebsiella pneumoniae consistently ranked among the top three gram-negative bacteria detected in hospitals, with an average detection rate of approximately 20% from 2015 to 2024. During this period, a total of 6,676 strains of K. pneumoniae were identified, including 714 strains classified as CRKP isolates, predominantly originating from sputum samples (58.10%), followed by urine samples (12.30%) and pus samples (9.40%).
4.2. Drug Resistance of <i>Klebsiella pneumoniae</i>
The detection rate of CRKP decreased from 9.77% in 2015 to 3.00% in 2016, but subsequently rose to 10.38% in 2024 (Table 1). The CRKP exhibits a higher resistance rate compared to carbapenem-sensitive K. pneumoniae (CSKP) (Table 2). The CSKP demonstrated a resistance rate of less than 30.00% for common antibiotics, except for cefoxitin and ampicillin/sulbactam, whereas CRKP exhibited a resistance rate of less than 50.00%, except for aminoglycoside antibiotics, and over 90.00% for β-lactam antibiotics. The resistance rates of levofloxacin and ciprofloxacin were notably high, at 91.60% and 92.90%, respectively.
| Year | KP | CRKP | CRKP Rate (%) |
|---|---|---|---|
| 2015 | 532 | 52 | 9.77 |
| 2016 | 538 | 16 | 3.00 |
| 2017 | 574 | 35 | 6.10 |
| 2018 | 607 | 57 | 9.39 |
| 2019 | 678 | 87 | 12.83 |
| 2020 | 627 | 99 | 15.79 |
| 2021 | 779 | 121 | 15.53 |
| 2022 | 704 | 76 | 10.79 |
| 2023 | 837 | 88 | 10.51 |
| 2024 | 800 | 83 | 10.38 |
Detection of Carbapenemase-Resistant Klebsiella pneumoniae Through the Years
| Antibiotics | CRKP (%) | CSKP (%) |
|---|---|---|
| Sulfamethoxazole/trimethoprim | 76.70 | 29.10 |
| Ceftazidime | 97.50 | 14.60 |
| Cefepime | 94.00 | 13.40 |
| Piperacillin-tazobactam | 99.00 | 7.10 |
| Amikacin | 25.60 | 5.60 |
| Cefoperazone/sulbactam | 97.30 | 2.10 |
| Levofloxacin | 91.60 | 9.60 |
| Cefuroxime | 99.90 | 23.10 |
| Ceftriaxone | 99.70 | 0.10 |
| Cefoxitin | 95.70 | 34.40 |
| Ertapenem | 97.40 | 9.60 |
| Ciprofloxacin | 92.90 | 22.60 |
| Gentamicin | 46.80 | 14.90 |
| Aztreonam | 98.40 | 15.20 |
| Ampicillin/sulbactam | 99.90 | 38.30 |
| Meropenem | 95.30 | 23.20 |
| Tobramycin | 41.50 | 0.40 |
Resistance Rates of Carbapenem-Resistant Klebsiella pneumonia and Carbapenem-Sensitive K. pneumoniae to Common Antibiotics
4.3. Risk Factors for Carbapenem-Resistant <i>Klebsiella pneumoniae</i>
The CRKP was more frequently isolated in Q1, Q2, and Q4 compared to Q3. Intensive care unit admission, hospitalization, and age over sixty were identified as clinical risk factors for CRKP infection. No significant difference was observed in the likelihood of CRKP infection between children aged three to nine years and those aged zero to two years. However, the probability of infection increased with age across other age groups, with the highest likelihood observed in individuals over sixty years old (Table 3).
| Risk Factor | CRKP/CSKP Ratio (%) | OR (95% CI) | P-Value |
|---|---|---|---|
| Specimen type | |||
| Blood | 36/505 (7.13) | 1 | NA |
| Sputum | 381/3282 (11.61) | 1.628 (1.143, 2.321) | 0.007 |
| Urine | 78/697 (11.19) | 1.570 (1.041, 2.368) | 0.032 |
| Type of patient | |||
| Outpatient | 7/284 (2.46) | 1 | NA |
| Inpatient | 534/5424 (9.85) | 3.994 (1.877, 8.499) | 0.000 |
| Age (y) | |||
| 0 - 2 | 7/396 (1.77) | 1 | NA |
| 3 - 9 | 1/56 (1.79) | 1.010 (0.122, 8.365) | 0.992 |
| 10 - 19 | 4/58 (6.89) | 3.901 (1.108, 13.741) | 0.034 |
| 20 - 39 | 25/419 (5.97) | 3.375 (1.444, 7.892) | 0.005 |
| 40 - 59 | 134/1568 (8.55) | 4.835 (2.243, 10.419) | 0.000 |
| > 60 | 370/3210 (11.53) | 6.521 (3.065, 13.874) | 0.000 |
| Quarter | |||
| Jul–Sep | 120/1854 (6.47) | 1 | NA |
| Jan–Mar | 137/1062 (12.90) | 1.993 (1.542, 2.576) | 0.000 |
| Apr–Jun | 154/1252 (12.30) | 1.897 (1.479, 2.434) | 0.000 |
| Oct–Dec | 130/1540 (8.44) | 1.304 (1.008, 1.687) | 0.043 |
| Ward | |||
| Non-ICU | 299/5299 (5.64) | 1 | NA |
| ICU | 124/409 (30.3) | 5.373 (4.259, 6.778) | 0.000 |
Analysis of Clinical Risk Factors for Carbapenem-Resistant Klebsiella pneumonia
4.4. Carbapenem-Resistant <i>Klebsiella pneumoniae</i> Drug Resistance Gene Analysis
Among the CRKP strains stored from 2020 to 2024, five strains were randomly selected each year and designated as 20K1-K5, 21K6-K10, 22K11-K15, 23K16-K20, and 24K21-K25, respectively. Their drug resistance genes and ST types were analyzed. The MLST type of all strains was ST11. Each strain contained the carbapenem resistance gene blaKPC-2, the ultra-broad spectrum β-lactamase resistance gene blaSHV, and the fosfomycin resistance gene fosA (except for 23K17). Additionally, they harbored various drug resistance genes, including ultra-broad spectrum β-lactamase resistance genes (blaTEM and blaCTX), aminoglycoside resistance genes (rmtB, ant, aac, aadA2), quinolone resistance genes (qnr, tetA, sul), chloramphenicol resistance gene (catA2), trimethoprim resistance gene (dfrA), and aminoglycoside resistance gene (aadA2) (Figure 1).
4.5. Fourier-Transform Infrared Spectroscopy Typing
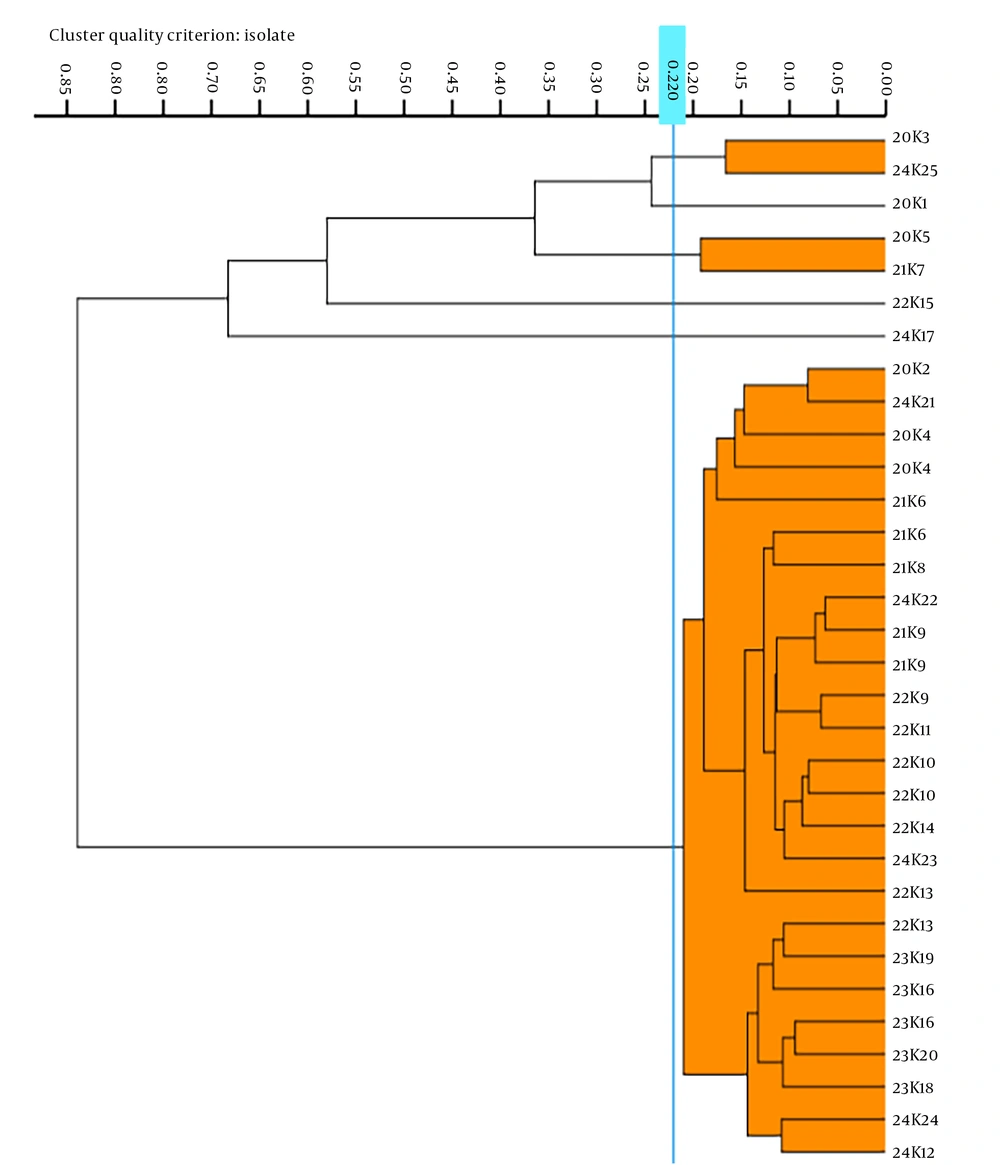
We conducted an analysis of 25 K. pneumoniae strains, selected from MH plates, using IRBT software, which automatically assigned a cut-off value of 0.220. The analysis categorized the 25 strains into six distinct types. Notably, strains 20K1, 22K15, and 23K17 formed unique clusters, while 20K5 and 21K7 were classified under the same type. Similarly, 20K3 and 24K25 were identified as belonging to the same type, and the remaining 14 strains shared another common type. The FTIR typing results indicated that the predominant strains circulating from 2020 to 2024 were clones of a single strain. Refer to Figure 2 for further details.
5. Discussion
In this study, we monitored the prevalence of CRKP in Lishui from 2015 to 2024. A total of 6 676 K. pneumoniae strains were identified, among which 714 were CRKP strains. While the proportion of ESBL-producing K. pneumoniae declined from 31.47% to 22.10% over the past five years (data not shown), the prevalence of CRKP initially declined from 9.77% in 2015 to a low of 3.00% in 2016, followed by a sharp increase, peaking at 10.38% in 2024. This trend contrasts with national CHINET (3) and Zhejiang province (12) data. A review of CRKP data from 2008 to 2014 revealed no cases at our hospital during 2008 - 2010; however, 18 strains (4.30%) were identified in 2011, rising to 52 strains (9.77%) in 2013, before dropping to 16 strains (3.00%) in 2016. Medical records indicate that the first CRKP detection at our hospital occurred in July 2011, following the transfer of a traumatic brain injury patient from a tertiary hospital in Shanghai.
The initial lack of infection control experience facilitated the rapid spread of CRKP within our intensive care unit (ICU). Following the patient’s discharge in August 2013, stringent containment measures were implemented, including a complete evacuation and decontamination of the ICU and the entire hospital, effectively curbing transmission within two years. As a result, the detection rate of CRKP in our ICU significantly decreased from 45.83% (44/96) in 2019 to 20.18% (23/114) in 2021, a trend that diverges notably from CHINET reports for China. This reduction may be attributed to the implementation of intestinal CRE screening and proactive interventions at our hospital starting in 2020 (13). These findings highlight the effectiveness of timely treatment and robust infection control measures in preventing the spread of CRKP.
The treatment options for CRKP infections remain severely limited, as cephalosporins, β-lactamase inhibitor combinations, fluoroquinolones, and aminoglycosides exhibit high resistance rates, rendering them unsuitable. However, the CRKP strains isolated in our hospital demonstrated significant sensitivity to tigecycline, polymyxin, and ceftazidime-avibactam. Despite the rise of antibiotic-resistant strains (14-16), these findings suggest potentially effective alternative therapies for managing CRKP infections in our hospital setting. The predominant drug resistance gene in CRKP isolates at our hospital is primarily blaKPC-2, along with various other resistance genes. The FTIR typing indicates that the primary epidemic strain from 2020 to 2024 has remained consistent within our hospital environment, underscoring the necessity for ongoing surveillance and stringent infection control measures to prevent potential CRKP clone spread and outbreaks.
Our findings align with a meta-analysis (17), which identified ICU admission as a significant risk factor for CRKP infection, likely due to the pathogen’s ability to persist on environmental surfaces, facilitating increased transmission among patients, ICU staff, and hospital surroundings (18, 19). Additionally, our data suggest that advanced age—particularly in patients over 60 years old—is associated with CRKP infections, potentially due to compromised immunity and underlying health conditions. Moreover, K. pneumoniae isolated from sputum exhibited a higher likelihood of carbapenem resistance compared to those from urine or blood, which may be attributed to colonization factors. Overall, our findings indicate an increasing trend in CRKP detection over the past decade. The blaKPC-2 gene remains the dominant drug-resistant determinant, with significant horizontal transmission of the same clone within our hospital, emphasizing the critical need for sustained infection control efforts.