1. Background
Escherichia coli, a member of the Enterobacteriaceae family, is one of the main causative agents of various infections in humans (1). Although E. coli is a normal component of the commensal human gut microbiota, several variants have evolved specialized pathogenic mechanisms that enable them to cause disease, either in the intestinal tract (diarrheagenic E. coli) or at extraintestinal sites (2, 3). Uropathogenic Escherichia coli (UPEC) is a subtype of extraintestinal pathogenic Escherichia coli (ExPEC) that originates from the commensal intestinal flora, colonizes the urinary tract, and causes urinary tract infections (UTIs) through a variety of virulence mechanisms (4-6).
β-lactam antibiotics are widely used to treat infections caused by E. coli (7-9). However, one of the main mechanisms of resistance to β-lactams is the production of β-lactamase enzymes, which hydrolyze the β-lactam ring, thereby inactivating the antibiotic (10). The most common β-lactamase enzymes include extended-spectrum β-lactamases (ESBLs) such as CTX-M, TEM, and SHV, which belong to Ambler class A, and plasmid-mediated AmpC β-lactamases (pAmpCs) such as CMY, DHA, and ACT, which belong to Ambler class C (11, 12). Among these, the CMY-type, particularly the CMY-2 variant, is the most prevalent pAmpC β-lactamase reported globally in Enterobacteriaceae, including E. coli (13-19).
The spread of ESBL and pAmpC β-lactamase-producing strains is largely due to the horizontal transfer of mobile genetic elements between pathogenic and non-pathogenic bacteria (20). Extended-spectrum β-lactamase-producing strains are resistant to third-generation cephalosporins (3GCs), such as ceftazidime and ceftriaxone (21, 22). In contrast, AmpC β-lactamases confer high-level resistance to 3GCs and cephamycins (e.g., cefoxitin and cefotetan), and are not inhibited by traditional β-lactamase inhibitors, thus limiting therapeutic options (23). To date, local studies in Iraq have largely ignored the prevalence of E. coli strains producing ESBL and CMY-2-type AmpC β-lactamases. Most recent reports have focused primarily on ESBL-producing E. coli isolates (24-26).
2. Objectives
This study aims to characterize the phenotypic and molecular features of ESBL-/CMY-2-type producing uropathogenic E. coli (pUPEC) isolates and to assign them to phylogenetic groups.
3. Methods
3.1. Isolation and Identification of Uropathogenic Escherichia coli Isolates
A cross-sectional study was conducted between November 2022 and March 2023 at the tertiary care unit of Al-Hillah Teaching Hospital in Al-Hillah, Babylon province, Iraq. A total of 150 non-duplicate E. coli isolates were recovered from both male and female inpatients diagnosed with UTIs (bacterial count ≥ 105 CFU/mL and white blood cell count ≥ 104 leukocytes/mL of urine). Escherichia coli isolates were detected by colony morphology on eosin methylene blue and MacConkey agar (HiMedia, India) after incubation at 37°C. The isolates were further confirmed as presumptive E. coli via VITEK® 2 Compact (bioMerieux, France) and biochemical tests (e.g., catalase, oxidase, methyl red/Voges-Proskauer, citrate and urease, triple sugar iron agar, and sulfide indole motility). The E. coli isolates were stored in brain heart infusion broth (HiMedia, India) with 30% glycerol at -20°C for further analysis (27).
3.2. Phenotypic Detection of Extended-Spectrum β-Lactamase and AmpC Production
Antibiotic susceptibility to cephalosporins, including ceftazidime (30 μg, 3GC), cefotaxime (30 μg, 3GC), and cefoxitin (30 μg, cephamycin) (Biomaxima, Poland), was detected via the disk diffusion method in accordance with the CLSI (28). All 3GC-resistant isolates were investigated for ESBL production via the combination disk test (CDT). Cefotaxime and ceftazidime (30 µg) disks, with and without clavulanic acid (10 µg) (Biomaxima, Poland), were used. Compared with antibiotics alone, UPEC isolates with ≥ 5 mm in diameter in the inhibition zones in the presence of cefotaxime/clavulanic acid and ceftazidime/clavulanic acid were considered indicative of ESBL-pUPEC isolates. A CDT, as described by Peter-Getzlaff et al. (29), was performed on all UPEC isolates presumed to be AmpC producers. In this test, a cefoxitin disk (30 µg) alone or in combination with cloxacillin (230 μg) (Biomaxima, Poland) was used. An increase of ≥ 4 mm in the inhibition zones of the cefoxitin/cloxacillin and cefoxitin disks was considered positive for AmpC production.
3.3. DNA Extraction and Detection of the Extended-Spectrum β-Lactamase and AmpC Genes
Total genomic DNA was extracted using the FavorPrep Genomic DNA Mini Kit (Favorgen, Taiwan) following the manufacturer's instructions. The DNA was used as a template for polymerase chain reaction (PCR) analysis. Uniplex-PCR was subsequently performed using previously described primers for blaCTX-M, blaSHV, blaTEM, and blaCMY-2 (Table 1) (30, 31). The total reaction mixture (20 μL) consisted of 8 μL of master mix, 7.5 μL of MgCl2, 0.5 μL of DNase/RNase-free sterile water, 2 μL of DNA template (50 ng), and 1 μL of each primer (Cyntol, Russia).
| Gene; Primer | Sequence (5′-3′) | PCR Conditions | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| blaCTX-M | 95°C, 1 min; 50°C, 1.5 min; 72°C, 1.5 min | 599 | (31) | |
| CTX-F | ACAGCAGATAATTCGCAA | |||
| CTX-R | AACCAGATCACCGCGATA | |||
| blaTEM | 95°C, 1 min; 60°C, 1.5 min; 72°C, 1.5 min | 500 | (31) | |
| TEM-F | TCAACAGCGGTAAGATCCTTGA | |||
| TEM-R | TGCAACTTTATCCGCCTCCA | |||
| blaSHV | 95°C, 1 min; 60°C, 1.5 min; 72°C, 1.5 min | 481 | (31) | |
| SHV-F | AAATGGATCTGGCCAGCG | |||
| SHV-R | AGCAGCTGCCGTTGCGAA | |||
| blaCMY | 95°C, 1 min; 55°C, 1.5 min; 72°C, 1.5 min | 1226 | (30) | |
| CMY-F | AACACACTGATTGCGTCTGAC | |||
| CMY-R | CTGGGCCTCATCGTCAGTTA |
Abbreviation: PCR, polymerase chain reaction.
3.4. Phylogenetic Group Analysis
The phylogenetic group of each UPEC isolate was detected according to the method described by Clermont et al. (32) using triplex PCR of the genes chuA and yjaA and the DNA fragment TSPE4.C2.
3.5. Sequencing
The amplified targets were sequenced via an automatic sequencer (Bioneer Corporation, South Korea). Nucleotide sequence alignment and analyses were performed online via the Basic Local Alignment Search Tool (BLASTn) program, which is available through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST).
4. Results
4.1. Isolation and Identification of Extended-Spectrum β-Lactamase-/AmpC-Producing Uropathogenic Escherichia coli Isolates
In this study, a total of 150 non-duplicate UPEC isolates were purified and identified using standard bacteriological methods. The CDTs for phenotypic detection of ESBL and AmpC production revealed that 134 isolates (89.3%) exhibited an ESBL phenotype, 103 isolates (68%) exhibited an AmpC phenotype, and 103 isolates (68%) showed a mixed ESBL/AmpC coproduction phenotype (Appendix 1 in Supplementary File). A total of 100 ESBL- and/or AmpC-positive isolates were selected for genotypic analysis.
4.2. Molecular Detection of Extended-Spectrum β-Lactamase- and CMY-2-Encoding Genes
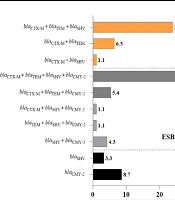
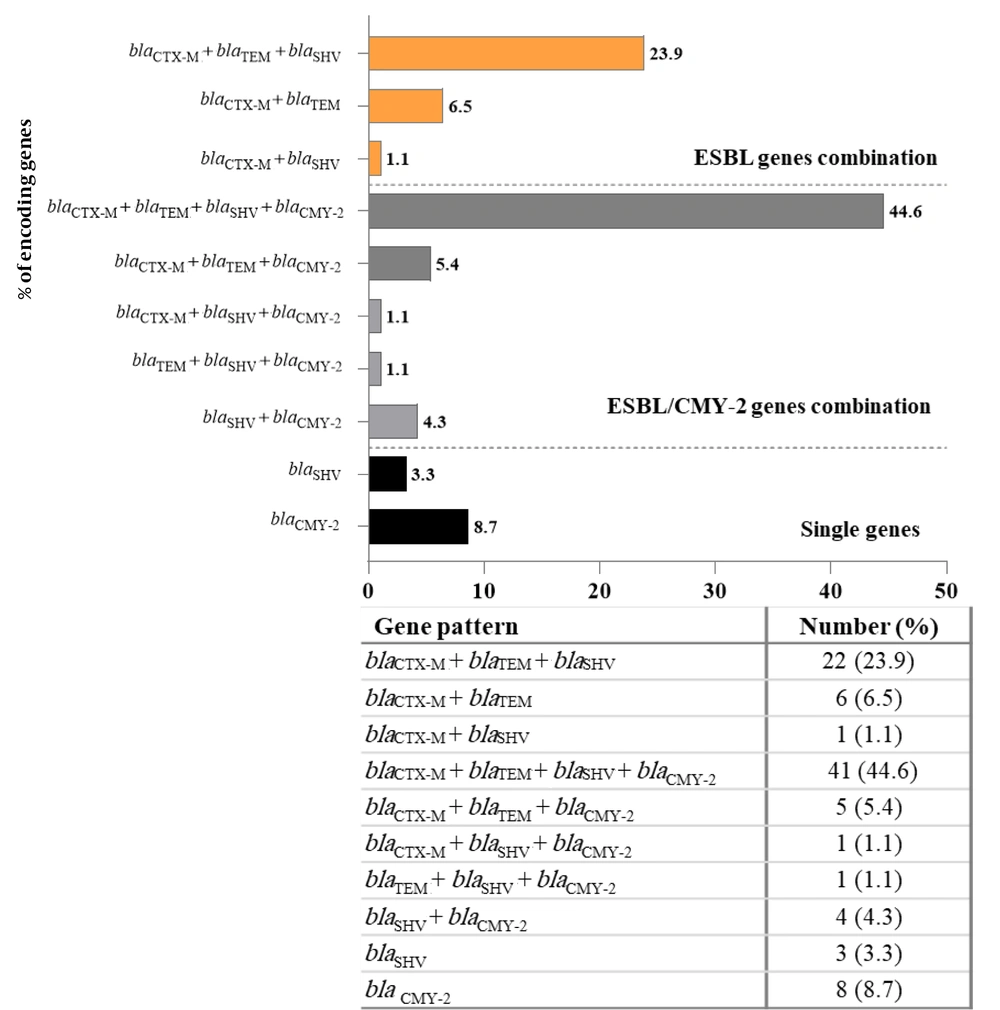
Approximately 84% (84/100) of the UPEC isolates were positive for ESBL-encoding genes. Among these, 76% (76/100), 75% (75/100), and 73% (73/100) were positive for the blaCTX-M, blaTEM, and blaSHV genes, respectively (Appendices 2 and 3 in Supplementary File). The PCR results revealed a prevalence of 60% for blaCMY-2 among the 100 isolates identified as AmpC-positive by the CDT method (Appendices 3 and 4 in Supplementary File). The PCR analysis of the 100 isolates demonstrated that 92% of UPEC isolates were positive for ESBL- and ESBL-/or CMY-2-type AmpC β-lactamase-encoding genes. Among the 92 isolates, 81 (88%) harbored more than one ESBL- and ESBL-/or CMY-2-type encoding gene, whereas only 11 (12%) of the isolates harbored only a single ESBL- or CMY-2-type encoding gene (Figure 1).
4.3. Distribution of Phylogroups Among the Extended-Spectrum β-Lactamase- and Extended-Spectrum β-Lactamase/CMY-2 Isolates
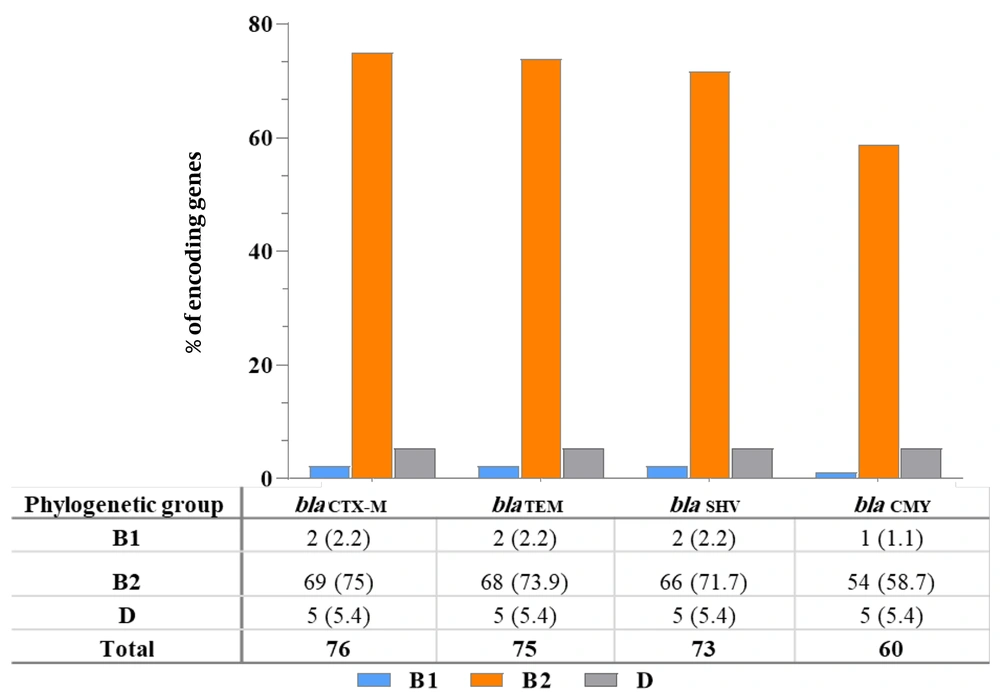
A total of 92 genotypically confirmed ESBL- and ESBL-/or CMY-2-positive isolates were assigned to phylogenetic groups. Most of the isolates were in group B2 (91.3%; 84/92), followed by group D (6.5%; 6/92) and group B1 (2.2%; 2/92), whereas none of the ESBL- and ESBL-/or CMY-2-type-pUPEC isolates were affiliated with group A (Table 2). The blaCTX-M (75%; 69/92), blaTEM (73.9%; 68/92), blaSHV (71.7%; 66/92), and blaCMY-2 (58.7%; 54/92) genes were most frequently detected in the extraintestinal phylogenetic group B2 (Figure 2).
| Phylogenetic Group | ESBL | CMY-2 | ESBL/CMY-2 | No. of Isolates |
|---|---|---|---|---|
| A | 0 | 0 | 0 | 0 |
| B1 | 1 (3.1) | 1 (1.9) | 2 (2.2) | |
| B2 | 30 (93.8) | 8 (100) | 46 (88.5) | 84 (91.3) |
| D | 1 (3.1) | 5 (9.6) | 6 (6.5) | |
| Total No. of isolates | 32 | 8 | 52 | 92 |
Abbreviation: ESBL, extended-spectrum β-lactamase.
a Values are expressed as No. (%).
5. Discussion
The UTIs have become a serious epidemiological threat, particularly in the treatment and control of severe infections caused by β-lactamase-producing E. coli isolates. This cross-sectional study describes the phenotypic and molecular characterization of UPEC isolates with reduced susceptibility to 3GCs and cefoxitin. Among the 150 UPEC isolates, 86.7% and 68.7% exhibited ESBL and AmpC phenotypes, respectively, which aligns closely with data reported from Wasit province, Iraq, where the prevalence of ESBL and AmpC phenotypes was 80.2% and 77%, respectively (33). The spread of these resistant strains is likely driven by poor antibiotic stewardship, misuse of antibiotics, and unregulated antimicrobial sales. Additionally, the influx of refugees may have increased selection pressure, facilitating the spread of multidrug resistance genes.
Confirmatory nucleic acid amplification remains essential for detecting ESBL- and CMY-2-type AmpC producers. In this study, 84% (84/100) of UPEC isolates carried at least one ESBL gene. blaCTX-M was most prevalent (90.5%; 76/84), followed by blaTEM (89.3%; 75/84) and blaSHV (86.9%; 73/84). Sequencing of three PCR amplicons identified blaCTX-M-15 (GenBank AY044436) as the dominant variant. Globally, CTX-M-15 is one of the most prevalent ESBL genotypes and is widely detected in E. coli isolates. This genotype has been reported in various systemic infections, including UTIs (34-36). Our findings are consistent with previous studies conducted in countries such as Lebanon, Saudi Arabia, Thailand, and China, which highlight CTX-M genes as the predominant ESBL determinants (37-40).
Chromosomal AmpC β-lactamase is not typically associated with increased enzyme production (29, 41). Therefore, the AmpC phenotype observed in cefoxitin-resistant UPEC isolates likely resulted from plasmid-mediated AmpC production, confirmed by PCR detection of the blaCMY-2 gene in 60% (60/100) of isolates (Appendix 4 in Supplementary File) Consistent with our findings, studies from Iran (42), Egypt (43), and New Zealand (44) reported CMY-2 prevalence rates of 72.4%, 86.9%, and 88%, respectively. The blaCMY-2 gene often coexists with other resistance genes on mobile genetic elements, contributing to multidrug resistance (18). In this study, co-occurrence of ESBL and CMY-2 genes was detected in 56.5% (52/92) of isolates, with 41 (44.6%) harboring blaCTX-M+blaTEM+blaSHV+blaCMY-2. Co-production of ESBL and CMY-2 β-lactamases has also been widely reported in countries such as Mexico, Europe, Iran, Sri Lanka, and Pakistan (45-49).
Escherichia coli isolates are classified into four phylogenetic groups: A, B1, B2, and D (32). The ExPEC strains causing UTIs are primarily found in groups B2 and D, while commensal strains are linked to A and B1 (2). In this study, blaCTX-M, blaTEM, blaSHV, and blaCMY-2 genes were most frequently detected in group B2, which accounted for 91.3% (84/92) of ESBL and/or CMY-2-pUPEC isolates, followed by group D (6.5%; 6/92). This high prevalence in B2 suggests an enhanced ability of ExPEC strains to acquire resistance genes via plasmid-mediated horizontal transfer, contributing to their spread (38, 50). Implementing proper infection control measures is essential to limit transmission. Routine susceptibility testing for 3GCs and cephamycins may support early detection and guide effective treatment.
This study was limited by its single-center design, small sample size, and focus on selected resistance genes without q-RT-PCR validation. Future research should involve larger, multicenter cohorts and broader molecular analysis to better understand resistance mechanisms in ESBL- and AmpC-producing E. coli.
5.1. Conclusions
The ESBL- and CMY-2-type isolates were highly prevalent in phylogroup B2, which includes ExPEC strains. These strains may cause UTIs and spread resistance genes via plasmid-mediated transfer. The findings underscore the urgent need for surveillance and resistance profiling in Iraqi hospitals to control dissemination and guide effective treatment strategies.