1. Background
The global health crisis caused by SARS-CoV-2 has underscored the urgent need for diagnostic tools that are not only sensitive and accurate but also rapid, accessible, and scalable, particularly in resource-limited settings (1-7). Although reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is widely regarded as the gold standard for COVID-19 detection due to its high sensitivity and specificity, its dependence on sophisticated laboratory infrastructure, skilled personnel, and lengthy processing times limits its practicality in many settings (8-14). These limitations have led to the exploration of alternative diagnostic platforms that can offer comparable accuracy with greater operational simplicity and speed (15, 16).
Loop-mediated isothermal amplification (LAMP) has emerged as one such alternative. Unlike PCR, which relies on thermal cycling to amplify DNA or RNA, LAMP performs amplification at a constant temperature (typically 60 - 65°C) using a DNA polymerase with strand displacement activity. It employs four to six primers that recognize six distinct regions of the target gene, enabling rapid and highly specific amplification, often producing detectable results within an hour (17-20). The method's high specificity, using six primers targeting unique sequences, reduces false positives and supports applications ranging from clinical diagnostics to environmental monitoring (20-24). Advances in LAMP's sensitivity have enabled the detection of low viral loads, critical for the early containment of infections (25). Additionally, the integration of real-time data through mobile health platforms enhances its utility in facilitating immediate public health responses (26).
Reverse transcription loop-mediated isothermal amplification (RT-LAMP), which incorporates reverse transcription, enables direct RNA sequence detection, making it highly effective for diagnosing RNA viruses such as SARS-CoV-2. Because LAMP requires only a simple heating device rather than a thermocycler, it is especially suitable for point-of-care and field-based applications in low-resource environments. Additionally, RT-LAMP integrates a reverse transcription step, allowing for direct detection of RNA viruses such as SARS-CoV-2 (27-33). However, despite these advantages, LAMP faces certain challenges that limit its widespread adoption in clinical diagnostics. One of the most critical issues is the risk of cross-contamination due to the large volume of amplified product generated, which can lead to false positives if stringent precautions are not observed. This has hindered its routine use in clinical laboratories despite its potential.
To overcome these limitations, ongoing research has focused on optimizing assay components, including primer design, incubation conditions, and elimination of the RNA extraction step to improve sensitivity, reduce contamination risks, and simplify the diagnostic workflow. In particular, LAMP assays targeting the conserved nucleocapsid (N) and envelope (E) genes of SARS-CoV-2 have shown promise for stable and accessible detection, aiding broader deployment, especially in underserved regions (33).
2. Objectives
This study aimed to develop and optimize an RT-LAMP assay for the rapid, sensitive, and cost-effective detection of SARS-CoV-2. It evaluated the assay's performance by comparing it with RT-PCR and optimized key parameters for accuracy. The study also explored the potential of colorimetric detection for simplified, point-of-care testing in diverse healthcare settings.
3. Methods
3.1. Clinical Sample Collection
Clinical samples for the study were collected at Emam Reza Clinic in Arak, Iran. Nasopharyngeal swabs were taken from participants and placed in viral transport medium (VTM), which were then transported under cold chain conditions to the molecular diagnostic center, where they were stored at 4°C. Sample processing occurred under a biosafety laminar flow class II cabinet to ensure safety and containment, and the samples were inactivated using either a heat treatment or a lysis buffer. A total of 80 samples, with both positive and negative results and varying cycle threshold (CT) values, were selected for analysis. Viral RNA was extracted using the QIAamp DSP Virus Kit (Qiagen, Hilden, Germany) as per the manufacturer’s protocol, and RT-PCR tests were performed according to instructions from Sansure Biotech (Changsha, China).
3.2. Sample Collection Procedure
Nasopharyngeal swabs in a Universal Transport Medium (UTM) were processed for direct RT-LAMP assays by vortexing for homogenization. Proteinase K was added in different concentrations to improve viral RNA release. After brief centrifugation, the samples were incubated at room temperature for 15 minutes and then heated at 95°C for 5 minutes to disrupt the viral envelope and release genomic material, optimizing the preparation for molecular diagnostics.
3.3. Reverse Transcription Loop-Mediated Isothermal Amplification Assay
The RT-LAMP assays were performed in reaction volumes of 20 µL, either using isolated RNA or directly from swabs (swab-to-RT-LAMP assays). Master mixes were prepared at room temperature immediately before use, containing 10 µL of the WarmStart Colorimetric RT-LAMP 2X Master Mix (M1800, M1804, New England, Biolabs).
3.4. Optimization of Reverse Transcription Loop-Mediated Isothermal Amplification Assay Sensitivity and Accuracy
3.4.1. Sample Preparation
The performance of the RT-LAMP assay was optimized by refining sample preparation and reaction conditions to improve detection without RNA extraction. Input volumes of 50 µL, 100 µL, and 200 µL were assessed for sensitivity and specificity. Samples were vortexed for 15 seconds and centrifuged 2 - 3 times for uniformity, with sub-samples of 5 µL, 10 µL, and 15 µL taken from the top layer to capture the concentrated viral fraction. Direct assays were compared to tests using 1 µL of extracted RNA, demonstrating the effectiveness of RNA extraction-free detection.
3.4.2. Proteinase K Concentration
Proteinase K, at concentrations of 1 - 2.5 mg/mL, was used to enhance viral RNA yield by lysing viral particles and releasing RNA (34). Samples were incubated at 55°C for 15 - 30 minutes to degrade proteins and disrupt the viral envelope. To ensure compatibility with RT-LAMP, Proteinase K was inactivated by heating the samples at 95°C for 5 - 10 minutes, enabling reliable assay performance (32).
3.4.3. Primer Concentration
In this study, RT-LAMP primers were designed to target the envelope (E) and nucleocapsid (N) genes of SARS-CoV-2, following the designs of Zhang et al. (31) and Broughton et al. (32) (Table 1). The primers were synthesized by Gene Fanavaran (Tehran, Iran) using RPC purification at a scale of 40 nmol. Two 10 × primer master mixes were created for the RT-LAMP assay, focusing on the N and E genes. The sequences and concentrations of the oligonucleotides were optimized to enhance sensitivity and specificity while minimizing non-specific amplification, which is essential for efficient amplification and accurate detection of viral genetic material.
| Component | Sequence/Description | Volume (μL) |
|---|---|---|
| DETECTR N-gene F3 | AACACAAGCTTTCGGCAG | 2 |
| DETECTR N-gene B3 | GAAATTTGGATCTTTGTCATCC | 2 |
| DETECTR N-gene FIP | TGCGGCCAATGTTTGTAATCAGCCAAGGAAATTTTGGGGAC | 16 |
| DETECTR N-gene BIP | CGCATTGGCATGGAAGTCACTTTGATGGCACCTGTGTAG | 16 |
| DETECTR N-gene LF | TTCCTTGTCTGATTAGTTC | 4 |
| Water (nuclease-free) | - | |
| DETECTR N-gene LB | ACCTTCGGGAACGTGGTT | 4 |
| NEB E1-F3 | TGAGTACGAACTTATGTACTCAT | 2 |
| NEB E1-B3 | TTCAGATTTTTAACACGAGAGT | 2 |
| NEB E1-FIP | ACCACGAAAGCAAGAAAAAGAAGTTCGTTTCGGAAGAGACAG | 16 |
| NEB E1-BIP | TTGCTAGTTACACTAGCCATCCTTAGGTTTTACAAGACTCACGT | 16 |
| NEB E1-LB | GCGCTTCGATTGTGTGCGT | 4 |
| NEB E1-LF | CGCTATTAACTATTAACG | 4 |
| Water (nuclease-free) | - | 52 |
N-Gene Detection Primers and E-Gene Detection Primers
Each primer master mix contains primers specifically designed to target the nucleocapsid (N) and envelope (E) genes of SARS-CoV-2. The F3 and B3 primers initiate amplification, while the FIP and BIP inner primers enhance specificity. Additionally, LF and LB primers serve as binding sites to accelerate the amplification process. Nuclease-free water is included to ensure proper dilution and maintain optimal concentrations in the final reaction mixture. Depending on the experimental design, the primer mixes can be used individually or in combination to assess the efficiency of viral gene detection.
3.4.4. Guanidine Hydrochloride
Guanidine hydrochloride, a chaotropic agent, enhances RNA extraction and detection efficiency in RT-LAMP assays for SARS-CoV-2 by disrupting hydrogen bonds and denaturing nucleoproteins and the viral envelope. This facilitates the release of intact RNA while protecting it from nuclease-mediated degradation. By destabilizing protein and nucleic acid secondary structures, guanidine hydrochloride improves RNA stability and amplification sensitivity. Its incorporation into RT-LAMP protocols has been shown to significantly enhance assay sensitivity and specificity. A guanidine chloride solution prepared using NEB (64504) and adjusted to pH ~8 with potassium hydroxide was evaluated at varying concentrations to optimize its impact on assay performance.
3.4.5. Betaine
The inclusion of betaine in RT-LAMP assays has been studied for its role in enhancing viral RNA detection, particularly for SARS-CoV-2. As a well-known PCR enhancer, betaine improves assay sensitivity and specificity by stabilizing RNA molecules and preserving template integrity during amplification. It reduces RNA secondary structure formation, promoting better primer binding and enhancing enzyme performance, which ultimately increases the sensitivity of the assay (35).
3.4.6. WarmStart NEB Buffer
To evaluate the performance of RT-LAMP assays for detecting SARS-CoV-2, two NEB buffers were used: WarmStart Colorimetric LAMP 2X Master Mix (M1800) and WarmStart RT-LAMP Buffer (M1804). The RT-LAMP Buffer (M1804) incorporates uracil-DNA glycosylase (UDG), which reduces contamination risks by preventing carryover from previous amplifications.
3.4.7. Incubation Time
The RT-LAMP assays typically require incubation periods of 30 to 60 minutes at optimal temperatures (60 - 65°C) for efficient primer binding to target RNA or cDNA. Insufficient incubation time may result in incomplete primer attachment, leading to reduced sensitivity and false negatives, especially in low viral load samples. Extended incubation improves primer-template hybridization and amplification yield but can also increase the risk of non-specific amplification or degradation of amplified DNA, affecting assay specificity. To optimize incubation time, experiments were conducted with varying durations (20 - 45 minutes), with amplification results monitored to determine the minimum time required for reliable target RNA detection.
3.4.8. Incubation Temperature
Temperature is a critical factor in RT-LAMP assays, influencing primer-template interactions and polymerase activity. RT-LAMP reactions are conducted at a constant temperature, typically between 60–65°C, where optimal primer annealing occurs. Temperatures above this range weaken primer binding, reducing specificity and sensitivity, and may denature the template RNA, hindering proper primer annealing. Lower temperatures may improve initial primer binding but may not provide sufficient energy for efficient polymerase activity, slowing amplification and decreasing sensitivity. To optimize these conditions, temperatures ranging from 58°C to 68°C were tested in increments of 1–2°C, with results analyzed to identify the ideal temperature for efficient primer annealing and polymerase extension. This systematic testing of time and temperature combinations enabled the determination of optimal conditions for maximizing the sensitivity, specificity, and reliability of the RT-LAMP assay, particularly for SARS-CoV-2 detection.
3.5. Monitoring Reverse Transcription Loop-Mediated Isothermal Amplification Results
3.5.1. Colorimetric Detection with Phenol Red
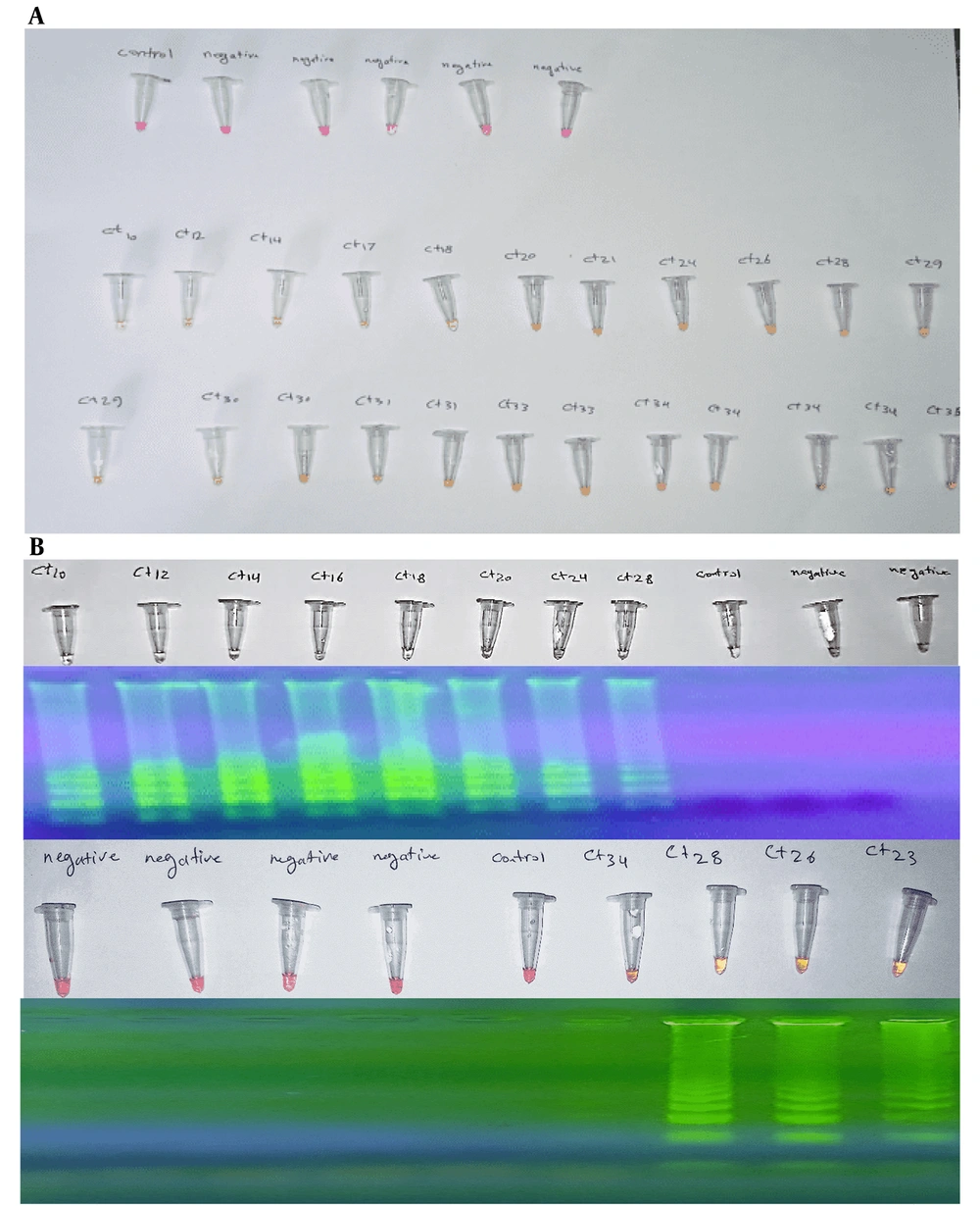
Phenol red serves as a pH indicator, undergoing a color change in response to the production of pyrophosphate during the RT-LAMP amplification process. As the reaction progresses, the increased pH resulting from DNA synthesis causes the solution to transition from red to yellow, indicating a successful amplification procedure. Following the RT-LAMP reaction, the reaction mixture was visually inspected for a color change. A red-to-yellow color shift confirmed positive amplification, providing a rapid, user-friendly, and equipment-free detection method.
3.5.2. Confirmation Through Gel Electrophoresis
Gel electrophoresis was employed to validate the results of colorimetric detection by confirming the presence and size of the amplified DNA products. This method offers a reliable secondary confirmation of RT-LAMP results. A 2% agarose gel with a safe DNA stain was prepared. Amplified RT-LAMP products, along with a DNA ladder for reference, were loaded into the gel. The gel was electrophoresed for 1 hour at 100 V. Bands were visualized under UV light. The band patterns were analyzed to confirm successful amplification, assessing both product presence and size.
3.5.3. Comparison with RT-PCR
RT-PCR, considered the gold standard for RNA detection, was used as a benchmark to compare the performance of the RT-LAMP assay. Samples tested in the RT-LAMP assay were reanalyzed using RT-PCR, following the manufacturer's protocol, with appropriate positive and negative controls included for validation. Threshold cycle (Cq) values were recorded to quantify viral RNA. Samples identified as positive by both RT-LAMP and RT-PCR were considered True Positives. Samples identified as negative by both assays were identified as True Negatives. Where the RT-LAMP assay provided a positive or negative result that was not corroborated by the RT-PCR, these were detected as False Positives/Negatives.
3.5.4. Performance Metrics
Performance metrics were calculated to evaluate the efficacy of RT-LAMP compared to RT-PCR.
1. Sensitivity: [True positives / (true positives + false negatives)]
2. Specificity: [True negatives / (true negatives + false positives)]
4. Results
4.1. Sample Preparation
The sample preparation methods were optimized to improve the reliability of downstream applications. A precise 100 µL of the viral sample was transferred into a sterile tube under controlled conditions to maintain sample integrity. The sample was initially vortexed vigorously for 5 seconds to resuspend settled particles and ensure homogeneity. A subsequent 15-second pulse vortexing step provided gentler mixing while preserving the sample. After vortexing, the sample was briefly allowed to stand, enabling the separation of the supernatant from particulates. A 5 µL aliquot of the supernatant was then pipetted for further analysis, minimizing contamination and ensuring consistency in subsequent steps. This optimized protocol significantly improved the efficiency of sample handling and contributed to the reproducibility of results in subsequent molecular assays.
4.2. Proteinase K Concentration
Specifically, 2 µL of Proteinase K was added to each sample vial. The vials were briefly vortexed for 2 seconds to ensure homogeneous mixing. Subsequently, a brief centrifugation was performed to collect the mixture at the bottom of the vial. The samples were incubated at room temperature for 15 minutes, followed by heating at 95°C for 5 minutes to disrupt the viral envelope and release genomic material. The samples were then immediately placed on ice to halt further reactions.
4.3. Primers
A comparative evaluation was conducted to test the effectiveness of using both sets of master primers simultaneously versus employing only the primers designed for the N gene. Results indicated that the N gene-specific primer set alone produced comparable outcomes to those obtained with both sets of primers. Therefore, subsequent experiments exclusively used the N gene master primer set. Increasing the concentrations of the forward loop primer (LF) and backward loop primer (LB) to 0.8 µM enhanced the efficiency of the RT-LAMP reaction, optimizing the overall assay performance.
4.4. Guanidine Hydrochloride
The addition of 40 mM guanidine hydrochloride to the RT-LAMP reaction significantly improved the assay's sensitivity, achieving up to a two-fold increase compared to reactions conducted without guanidine hydrochloride.
4.5. Betaine
The introduction of betaine into the RT-LAMP reaction did not yield any positive results. This suggests that betaine may not enhance the assay's sensitivity or effectiveness in detecting the target nucleic acid.
4.6. WarmStart NEB Buffer
The use of WarmStart NEB buffer containing UDG (Uracil-DNA Glycosylase, M1804) markedly reduced the incidence of false positives in the RT-LAMP assay. This improvement highlights the utility of UDG in enhancing the assay's specificity and reliability by mitigating contamination risks from previously amplified products.
4.7. Incubation Conditions
Extending the final incubation time to 40 minutes at 62°C yielded optimal reaction efficiency. This extended duration likely facilitated a more complete amplification of the target nucleic acid, thereby improving the sensitivity and reliability of the assay.
4.8. Final Optimized Protocol
A 10 µL aliquot of Colorimetric LAMP buffer (M1804) was prepared, and 2 µL of an optimized primer mixture was added, consisting of 0.8 µM LF and LB primers, 0.2 µM F3 and B3 primers, and 1.6 µM FIP and BIP primers. To enhance assay sensitivity, 40 mM Guanidine Hydrochloride was incorporated. The samples were heated at 95°C and then immediately placed on ice to stabilize viral RNA and quickly reduce the reaction temperature. The final reaction volume was adjusted to 20 µL with nuclease-free water (NEB B1500). A negative control was prepared by replacing 2 µL of the sample with nuclease-free water. The reaction mixture was incubated at 62°C for 40 minutes. A phenolic dye in the buffer enabled visual result interpretation: Negative samples appeared pink, while positive samples turned yellow or orange. Gel electrophoresis confirmed the presence of amplified products in positive samples (Figure 1).
| Step | Optimized Conditions |
|---|---|
| Sample preparation | 100 µL of viral sample transferred to a sterile tube, vortexed for 5 seconds, followed by 15-second pulse vortexing. A 5 µL aliquot of the supernatant was taken for analysis. |
| Proteinase K concentration | 2 µL of Proteinase K added to each sample, vortexed for 2 seconds, centrifuged briefly, incubated at room temperature for 15 minutes, then heated at 95°C for 5 minutes. Samples placed on ice to stop reactions. |
| Primers | N gene-specific primer set used. The LF and LB concentrations increased to 0.8 µM to enhance assay efficiency. |
| Guanidine hydrochloride | 40 mM guanidine hydrochloride added to the RT-LAMP reaction, improving sensitivity and increasing assay performance up to two-fold. |
| Betaine | No positive results observed with the addition of betaine, indicating it does not enhance sensitivity. |
| WarmStart NEB buffer | WarmStart NEB buffer containing UDG (M1804) used to reduce false positives by mitigating contamination risks from amplified products. |
| Incubation conditions | Final incubation time of 40 minutes at 62°C to maximize reaction efficiency and improve sensitivity. |
| Final optimized protocol | 10 µL Colorimetric LAMP buffer + 2 µL primer mixture (0.8 µM LF and LB primers, 0.2 µM F3 and B3 primers, 1.6 µM FIP and BIP primers). 40 mM Guanidine Hydrochloride added. Reaction mixture incubated at 62°C for 40 minutes. Visual result interpretation with phenolic dye (pink for negative, yellow/orange for positive). Gel electrophoresis confirms amplification. |
Summary of Optimized Conditions for Reverse Transcription Loop-Mediated Isothermal Amplification Assay
4.9. Comparison with RT-PCR
A comparative evaluation of the optimized RT-LAMP assay and RT-PCR for COVID-19 detection revealed that RT-LAMP demonstrated comparable sensitivity to RT-PCR, successfully detecting viral RNA in samples with a CT value up to 35. This performance highlights RT-LAMP's ability to amplify and detect low viral RNA levels, making it a viable alternative to RT-PCR. Additionally, RT-LAMP offers a significantly faster turnaround time, making it ideal for point-of-care testing, where timely results are crucial. Its ability to detect viral RNA with a CT value of up to 35 emphasizes its efficacy in detecting infections, particularly in the early stages with lower viral loads.
4.9.1. Direct Use of Pharyngeal Swab Samples
The data are presented in a summarized format in Table 3.
| Category | Count | Total |
|---|---|---|
| TPs | 51 | |
| CT < 24 | 18 | |
| CT 24 - 30 | 22 | |
| CT > 30 | 11 | |
| FNs | 10 | |
| CT 24 - 30 | 0 | |
| CT > 30 | 10 | |
| TNs | ||
| Negative | 13 | 13 |
| FPs | 2 | 2 |
Summary of Reverse Transcription Loop-Mediated Isothermal Amplification Assay Performance
4.9.2. Use of Extracted RNA
The data are presented in a summarized format in Table 4.
| Category | Count | Total |
|---|---|---|
| TPs | ||
| CT < 24 | 18 | 52 |
| CT 24 - 30 | 22 | |
| CT > 30 | 12 | |
| FNs | ||
| CT 24 - 30 | 0 | 9 |
| CT > 30 |
Summary of Reverse Transcription Loop-Mediated Isothermal Amplification Assay Performance
4.9.3. Performance Metrics
Table 4 presents the performance metrics of the RT-LAMP assay for viral RNA detection, comparing the results from directly using pharyngeal swab samples with those from extracted RNA. Sensitivity measures the assay's ability to correctly identify positive cases, while specificity evaluates its accuracy in excluding false positives. Efficiency reflects the overall proportion of correctly identified cases, combining true positives and true negatives.
| Metric | Calculation | Pharyngeal Swab (%) | Extracted RNA (%) |
|---|---|---|---|
| Sensitivity | True positives/(true positives + false negatives) | ≈ 83.61 | ≈ 85.25 |
| Specificity | True negatives/(true negatives + false positives) | ≈ 86.67 | 100 |
| Efficiency | (True positives + true negatives)/total samples | ≈ 84.21 | ≈ 88.16 |
Performance Metrics Results
5. Discussion
The RT-LAMP assay demonstrated a sensitivity of 83.61% and an overall efficiency of 84.21%. While this performance is promising, it falls short when compared to the sensitivity of traditional RT-PCR assays, which often exceed 90%. The observed sensitivity issues, particularly with samples showing high CT values (CT > 30), point to a significant limitation of the RT-LAMP assay: The risk of false negatives in samples with low viral loads. This sensitivity gap highlights a key challenge in the optimization of LAMP tests, particularly in the detection of SARS-CoV-2, where early detection is crucial for controlling transmission.
The failure to achieve a sensitivity level comparable to RT-PCR is likely due to several factors, including the need for further optimization of the primer design, reaction conditions, and detection reagents. In our study, while we focused on improving sensitivity by optimizing key parameters such as primer concentrations, incubation time, and temperature, the overall sensitivity remained lower than desired. Although the use of colorimetric detection offered significant advantages in terms of accessibility and ease of use, this method does not address the inherent limitations of LAMP in terms of sensitivity for low viral load detection.
Furthermore, the incorporation of direct swabs without RNA extraction, while simplifying the process and making it more suitable for point-of-care settings, may also have contributed to reduced sensitivity, as the sample preparation and viral RNA extraction process in RT-PCR often leads to more concentrated and pure genetic material for detection. Comparative results from studies by Yan et al. (36) and Subali and Wiyono (37) showed similar performance to our study, with RT-LAMP achieving around 93% sensitivity, aligning with existing findings in SARS-CoV-2 diagnostics. The slight differences observed could be attributed to the variations in RNA extraction methods, primer designs, and sample handling.
Despite the lower sensitivity compared to RT-PCR, RT-LAMP still offers considerable potential, particularly in resource-limited settings where the infrastructure for traditional PCR testing may not be available. Optimizing primer design and concentration, as demonstrated by Sarwan et al. (38), significantly improved sensitivity and enabled reliable detection even at low viral loads, which is particularly beneficial for point-of-care applications. This work also builds on the original LAMP findings by Notomi et al. (39), emphasizing the value of visual result interpretation, which makes RT-LAMP an effective diagnostic tool in rural or low-resource settings where laboratory infrastructure is limited.
This study successfully developed and optimized an RT-LAMP assay for the detection of SARS-CoV-2, offering a promising alternative to traditional RT-PCR methods. The RT-LAMP assay has several advantages, including a simplified workflow that eliminates the need for complex thermal cycling, rapid results (under one hour), and colorimetric detection for easy visual interpretation. The optimized protocol demonstrated strong sensitivity and specificity, especially for extracted RNA from pharyngeal swabs, making it reliable for various diagnostic settings.
While the RT-LAMP assay offers rapid results within one hour and requires minimal equipment, making it an ideal candidate for use in field and point-of-care settings, its current sensitivity limitations need to be addressed for it to become a truly reliable alternative to RT-PCR.
5.1. Conclusions
In conclusion, while the optimized RT-LAMP assay represents a significant advancement in molecular diagnostics, more research is needed to improve its sensitivity, especially for low viral loads, to fully compete with the gold-standard RT-PCR method. Additionally, integrating RT-LAMP with mobile health technologies could enhance its utility for real-time data collection, surveillance, and outbreak management, particularly in underserved regions. Continued innovation in assay optimization will be crucial for improving global health diagnostics and enhancing preparedness for future infectious disease outbreaks.