1. Background
White spot syndrome virus (WSSV) is a highly virulent pathogen affecting cultured shrimp, causing up to 100% mortality in 3 - 10 days (1). It is a large, enveloped double-stranded DNA virus belonging to the Whispovirus genus of the Nimaviridae family, primarily affecting marine shrimp species (2). The WSSV was first detected in Taiwan in 1992 and has since spread globally (3). The dUTPase enzyme plays a crucial role in nucleotide metabolism by hydrolyzing dUTP to dUMP, preventing its misincorporation into DNA. Both shrimp and WSSV dUTPases have similar functions but differ in structural characteristics (Appendix 1 in Supplementary File, 14). These enzymes have been proposed as potential drug targets for treating infectious diseases (4-6).
Phytochemicals from plants like Cyperus rotundus and Salvia rosmarinus have shown potential in combating viral diseases. Cyperus rotundus has antiviral properties against several viruses, including hepatitis A (7, 8). Salvia rosmarinus contains bioactive compounds with antioxidant and anticancer effects (9, 10). Computational approaches, such as molecular docking and molecular dynamics (MD) simulations, are enhancing drug discovery by evaluating ligand stability and interactions with target enzymes like dUTPase (11). These methods are efficient, cost-effective, and less time-consuming than traditional methods (11).
2. Objectives
In the present study, we examined the dUTPase enzyme as a potential path to interfere with WSSV and explored the potential of phytochemicals from C. rotundus and S. rosmarinus in combating shrimp WSSV using molecular docking.
3. Methods
3.1. 3D Protein Crystal Structure
The WSSV dUTPase 3D structure was obtained in PDB format from the Protein Data Bank (PDB). The Discovery Studio Visualizer software package was used to prepare the protein, eliminating all heteroatoms and water molecules from the structure (12).
3.2. Phytochemical Selection and Preparation
Phytochemicals present in C. rotundus and S. rosmarinus were reviewed in the literature, identifying constituents from the Indian Medicinal Plant, Phytochemistry and Therapeutics (IMPPAT) database (13).
3.3. Ligand Preparation
To predict binding sites, LIGPLOT analysis was conducted using the PDBsum program v4.5.3. The input is a four-digit PDB code for dUTPase (5Y5P) (14).
3.4. Protein Preparation and Validation
The Ramachandran plot (RP) was applied using the SAVES program v6.1 to evaluate the precision of the predicted protein structure (14). The quality of the model protein structure was then validated using ERRAT (15).
3.5. Screening of Compounds
3.5.1. Absorption, Distribution, Metabolism, and Excretion Analysis
Absorption, distribution, metabolism, and excretion (ADME) defines the key pharmacokinetic parameters that influence the bioavailability and efficacy of a drug compound. The SwissADME server (16) and pkCSM online tool (17) were employed to determine the different physicochemical descriptors and to predict various ADME parameters, pharmacokinetic properties, and drug-like characteristics.
3.5.2. Toxicity Analysis
Prediction of the toxicity of compounds offers valuable insights into the possible risks associated with a drug. In this study, Protox_II and pkCSM were used to investigate the toxicity of the selected compounds (18).
3.5.3. Lipinski Rule Analysis
The Lipinski rule, also known as the rule of five, was used to assess the oral bioavailability of a compound using data from the SwissADME server (18).
3.6. Molecular Docking
Molecular docking analyzes the binding affinity and nature of the interaction between a selected ligand and protein. Molecular docking simulations were conducted using PyRx software (18).
3.7. Analysis of Docked Complex and Visualization
Discovery Studio v4.1 was used to visualize the docking interaction and provide detailed insights into the 2D and 3D docked structure. This step is crucial for investigating and identifying nonbonding amino acid and ligand interactions (19).
3.8. Molecular Dynamic Simulation
Analysis of the thermodynamic stability of the receptor-ligand system was conducted using the GROMACS program. The root mean square deviation (RMSD), radius of gyration (Rg), root mean square fluctuation (RMSF), quantity of hydrogen bonds (H-bonds), salt bridges, and solvent accessible surface area (SASA) were calculated to monitor the stability of the MD simulations (20).
4. Results
4.1. Protein Structure
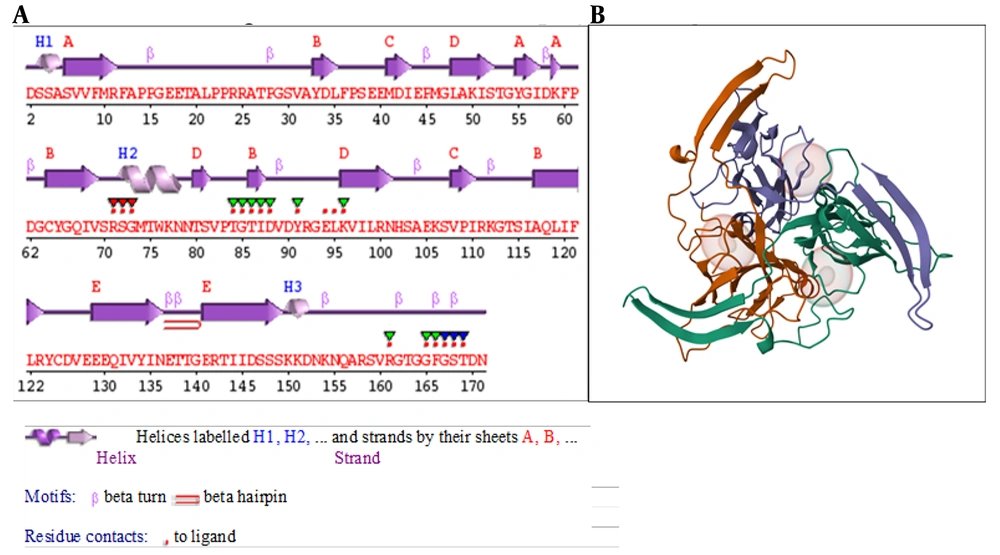
The secondary structure of the dUTPase protein (Figure 1A) is represented by α-helices and β-sheets coupled with different structural patterns like β-hairpins, and β- and γ-turns. Boxes around amino acid codes indicate catalytic residues. The red dots displayed below the figure, above the single-letter codes, indicate residues involved in interactions with the attached ligand(s), while colored lines below the codes indicate residues associated with PROSITE patterns. The intensity of the color indicates the degree of conservation of the residue within the pattern, with a deeper color indicating higher conservation. The 3D crystal structure of the dUTPase protein (PDB ID: 5y5p) was retrieved from the PDB as displayed in Figure 1B.
4.2. Protein Validation
4.2.1. Z-Score Plot
The ProSA-web server was utilized to assess the refined dUTPase protein quality. The findings indicated a Z-score of -4.03, which falls within the range of Z-scores observed for experimental native structures of comparable sizes.
4.2.2. Ramachandran Plot Analysis
The RP is a crucial tool for the validation of the protein model quality and accuracy through PROCHECK analysis. PROCHECK evaluates the stereochemical quality of protein structures, and the RP specifically assesses the distribution of phi (ϕ) and psi (ψ) dihedral angles for amino acid residues. The total number of residues in the refined dUTPase protein was found in the most favored regions (97.2%, Appendix 2 in Supplementary File), whereas the non-refined dUTPase protein result was 93.0% of amino acid residues in the core region.
4.2.3. ERRAT Plot
The ERRAT plot is a tool commonly used for the validation of protein structures. A well-distributed and elevated ERRAT score provides confidence in the reliability of the protein model, reinforcing its suitability for subsequent analyses such as molecular docking. The ERRAT scores obtained in this study were 90.5% before and 97.3% after refinement (Appendix 3 in Supplementary File). Higher scores imply better agreement between the model and the expected high-quality structure.
4.2.4. Verify3D Analysis
This analysis revealed that a substantial portion of the residues, precisely 82.35%, exhibited an averaged 3D-1D score equal to or less than 0.1. This outcome signifies a high degree of agreement between the model 3D structure and its corresponding 1D amino acid sequence. A 3D-1D score below 0.1 suggests that most residues are in favorable spatial conformations, reinforcing the reliability of the protein model. This alignment between the predicted and expected structural features is indicative of a well-folded and accurately modeled protein, enhancing confidence in its suitability for subsequent computational analyses.
4.3. Ligand Properties
The interactions between proteins and ligands were produced using the LIGPLOT software, which facilitates the visualization and analysis of protein-ligand interactions. This tool was employed to assess the presence of hydrophobic interactions, other non-covalent interactions, and the number of H-bonds between the amino acid residues and ligand located within the active sites of the protein. The results identified the four interacting residues LYS96, SER72, PRO502, and ASP88. Generally, six H-bonds (blue line) were recognized.
4.4. Molecular Docking Simulation
Phytochemicals were identified in the literature and specific proteins were chosen. The optimal intermolecular interactions were then identified through molecular docking studies. A total of 28 compounds were applied to the molecular docking process using the PyRx tool AutoDock Vina wizard. Among the tested compounds, three compounds (two from C. rotundus and one from S. rosmarinus) displayed the highest inhibitory activity against the targeted protein with Vina docking scores of -9.3, -8.8, and -7.8 kcal/mol, respectively (Appendix 15 in Supplementary File).
4.5. Protein-Ligand Interaction Analysis
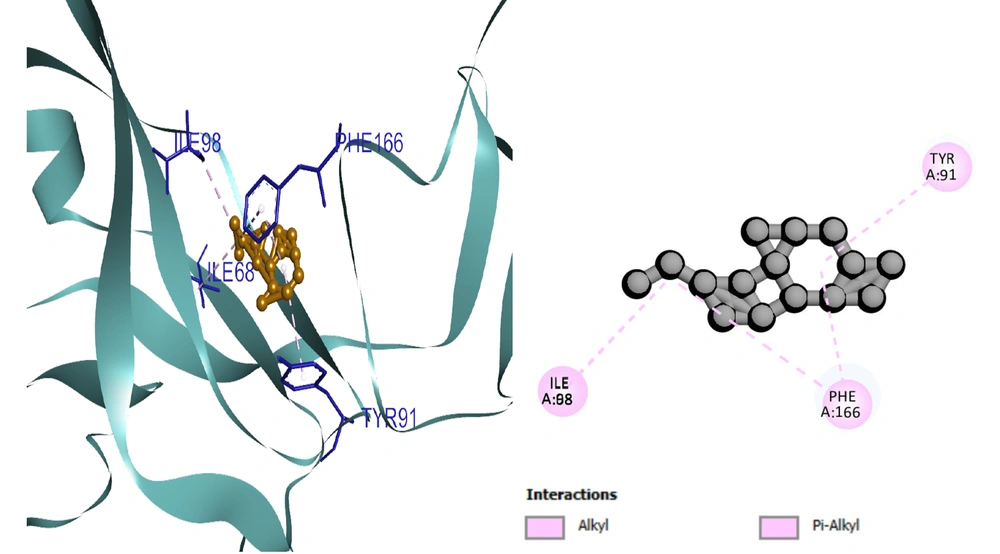
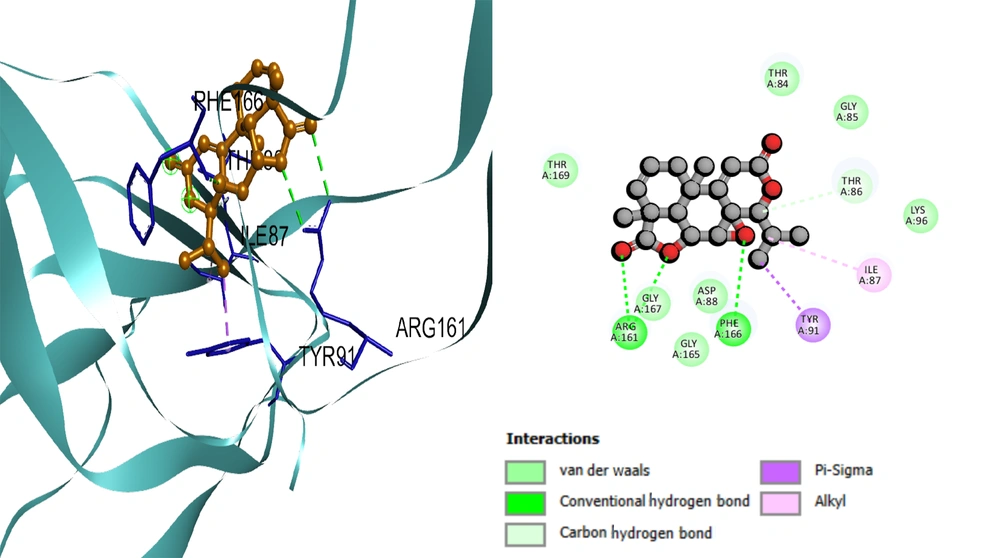
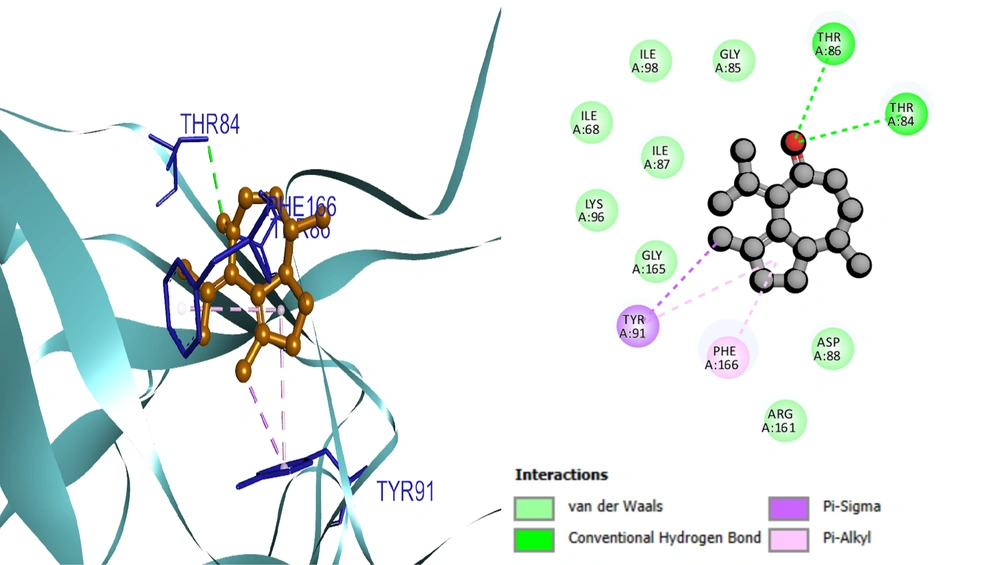
Based on the docking results, the protein-ligand interaction was visualized using the BIOVIA Discovery Studio Visualizer tool. The first compound, selinene (CID:442393), formed van der Waals (vdW), pi-sigma, alkyl, and pi-alkyl hydrophobic interactions with the four amino acid residues (ILE98, ILE68, PHE166, TYR91) situated at the pocket region (Figure 2). The second compound, podolide (CID:99535), forms four H-bonds in the THR169, ARG161, ASP94, and THR86 amino acid residue positions. Besides, three additional hydrophobic bonds were found at TYR91, PHE166, and THR86 amino acid residues (Figure 3). The third compound, zierone (CID:91752839), forms two conventional H-bonds at the THR86 and THR84 amino acid residue positions. Two additional bonds are found at TYR91 and PHE166 amino acid residues (Figure 4). All three compounds overlapped at PHE166 in the active site with the natural substrate.
4.6. Absorption, Distribution, Metabolism, and Excretion Analysis in Silico
The ADME analysis of all three compounds revealed remarkably favorable pharmacokinetic profiles across almost all evaluated parameters. The compounds showed favorable absorption with water solubility, Caco2 cell permeability, and gastrointestinal (GI) absorption. Besides, the three compounds showed P-glycoprotein I and II inhibitor activity and showed lower BBB and CNS permeability. The top three compounds were CYP1A2 inhibitors (Appendix 16 in Supplementary File). According to the Lipinski analysis, the three compounds do not violate the five rules.
4.7. Toxicity Analysis in Silico
Notably, the active cluster associated with selinene (CID:442393) appeared smaller than those of the other two compounds. However, most of the toxicity parameters of the three compounds were predominantly clustered within the inactive category, displaying a tendency towards inactive toxicity profiles, with probabilities equal to or greater than seven. More specifically, they showed a low toxic effect on organs including the liver, and were inactive for cytotoxicity, mutagenicity, carcinogenicity, and Tetrahymena pyriformis toxicity. Besides, they did not elicit a significant or adverse response to the eight-stress response pathways tested.
4.8. Molecular Dynamic Simulation
4.8.1. Root Mean Square Deviation
The resulting stability trends are different for each ligand, reflecting variability in the quality of their interaction with the target protein. Among the three ligands, 99535, represented by the green line (Appendix 4 in Supplementary File), has the lowest and most stable RMSD values, remaining below 0.25 nm during the simulation. Ligand 91752839 exhibits moderate binding. Ligand 442393, although initially exhibiting stability, undergoes significant deviations indicative of weak binding interactions. Therefore, these findings indicate ligand 99535 to be the optimal candidate for maintaining the structural integrity of the dUTPase protein.
4.8.2. Root Mean Square Fluctuation
The RMSF gives a quantitative measure of atomic displacement that reflects the stability or flexibility of individual residues. For ligand 442393, the structure is almost regular, having medium stability with local flexibility in terminal and loop regions. Ligand 91752839 is more flexible and, near residue 800, the interactions are not so stable. Ligand 99535 (Appendix 5 in Supplementary File) exhibits the highest stability with the lowest RMSF value reflecting strong binding to maintain the structure. Therefore, the superior stabilizing effect of ligand 99535 on the dUTPase protein reinforces the previous indication of its favorable binding characteristics.
4.8.3. Solvent Accessible Surface Area
Changes in SASA effectively explain structural rearrangements and the overall compactness of the protein-ligand complexes. Ligand 99535, represented by the green line (Appendix 6 in Supplementary File), shows the lowest and steady SASA in the range 410 - 420 nm2 throughout the simulation. This low fluctuation strongly indicates that this protein-ligand interaction is stable and compact. Minimal changes following exposure to solvent reflect well-maintained structural integrity favoring strong and reliable binding. In summary, ligand 99535 possesses a minimum fluctuation in SASA and a compact protein structure. The highest fluctuations were measured for ligand 442393, suggesting destabilization and poorer binding with the target receptor. Ligand 91752839, with moderate stability and some flexibility, is intermediate in nature. These findings confirm better stability and higher binding potential of ligand 99535.
4.8.4. Radius of Gyration
The radius of Rg gives a measure of the general distribution of the mass of the protein relative to its center. The lower the Rg, the more compact the protein. Ligand 99535 presents the highest compactness and stability, reflected in relatively constant Rg values (Appendix 7 in Supplementary File). Instead, ligands 442393 and 91752839 present fluctuation of higher values, showing lower stability and structural flexibility. These results support the previous analyses, indicating ligand 99535 to be the best candidate to maintain the integrity of the structure of the dUTPase protein.
4.8.5. Hydrogen Bond Analysis
The H-bonds are crucial for protein-ligand stability. Ligand 99535 consistently formed 3 - 4 H-bonds during simulations, indicating strong interactions. In contrast, ligand 442393 lacked H-bond donors or acceptors, suggesting hydrophobic or vdW interactions instead. The absence of H-bonds does not necessarily imply weaker binding; rather, it may involve alternative interaction mechanisms.
4.8.6. Interaction Energy Analysis
Ligand 99535 showed stable electrostatic interactions with Coulombic short-range (Coul-SR) energy around -80 kJ/mol and strong vdW interactions with Lennard Jones short-range (LJ-SR) energy around -120 kJ/mol (Appendix 8 in Supplementary File). This stability indicates a favorable binding environment, making ligand 99535 a strong candidate for drug development. Ligand 442393 had minimal electrostatic interactions but stable vdW interactions between -100 and -140 kJ/mol, indicating a binding mechanism involving hydrophobic forces (Appendix 9 in Supplementary File). Ligand 91752839 exhibited weak electrostatic and vdW interactions, reflecting less stable binding (Appendix 10 in Supplementary File).
4.8.7. Principal Component Analysis
Ligand 99535 displayed two well-separated clusters, indicating high stability and dynamic flexibility. Ligand 442393 showed a dispersed pattern, suggesting higher structural flexibility and weaker binding. Ligand 91752839 had a compact cluster, indicating limited flexibility and constrained binding. These results confirm that ligand 99535 has higher stability and adaptability, while ligand 442393 shows flexibility and ligand 91752839 exhibits restricted dynamics.
4.8.8. Secondary Structure Analysis
Ligand 99535 maintained stable α-helices and β-sheets throughout the simulation, indicating strong protein-ligand interaction. Ligand 442393 showed moderate fluctuations in secondary structures, suggesting increased flexibility. Ligand 91752839 had significant disturbances in secondary structures, reflecting less stable interactions and reduced structural integrity. Overall, ligand 99535 is the strongest candidate for maintaining protein structural integrity.
4.8.9. Binding Energy Decomposition
Ligand 99535 had a favorable enthalpy/entropy balance (ΔH ≈ -15 kcal/mol, -TΔS ≈ 3 kcal/mol), resulting in ΔG ≈ -12 kcal/mol (Appendix 11 in Supplementary File). This balance indicates strong interactions with minor structural reorganization. Ligand 442393 had strong enthalpic contributions but larger entropy losses, suggesting stable interactions with greater conformational changes (Appendix 12 in Supplementary File). Ligand 91752839 exhibited strong enthalpic forces with significant entropic penalties, indicating extensive structural rearrangements (Appendix 13 in Supplementary File). Overall, ligand 99535 is thermodynamically the most efficient binder due to its balanced contributions.
5. Discussion
This study examines computational methodologies used in antiviral drug discovery, specifically focusing on molecular docking and dynamics simulations for identifying phytochemical inhibitors against WSSV. The analysis incorporates findings from a comprehensive study that screened over 1,000 compounds and identified promising candidates through rigorous computational validation. The application of the specified Lipinski’s rule threshold (Appendix 17 in Supplementary File) excluded 90% of the phytochemicals, where 28 compounds were approved after toxicity criteria (Appendix 18 in Supplementary File).
The WSSV poses a significant threat to crustacean aquaculture, causing substantial economic losses since its identification in 1992. Despite this, no specific antiviral drugs are available to treat WSSV infections, highlighting a critical gap in disease control. Recently, plant-derived compounds have emerged as potential antiviral agents. Cyperus rotundus and S. rosmarinus have been studied for their medicinal properties, including antiviral activities against pathogens like the avian infectious bronchitis virus (IBV) (21-24). Computational methods such as molecular docking and MD simulations are used to explore the antiviral potential of these plant constituents against WSSV (25). These computational approaches predict binding affinities and inhibitory effects, helping identify promising candidates for experimental validation. Recent studies on C. rotundus-derived nanoparticles demonstrated antiviral efficacy against avian coronaviruses, reinforcing the plant’s therapeutic potential (21).
The structural properties of compounds from C. rotundus and S. rosmarinus are analyzed using resources like PubChem (13). Pharmacological reviews of Salvia species highlight their broad-spectrum antiviral mechanisms, including inhibition of viral entry and replication (24). Toxicity predictions are crucial for assessing potential risks and informing drug development decisions (26). For instance, compounds like β-amyrin and stigmasta-5,22-dien-3-ol from C. rotundus have shown promising binding interactions in studies against SARS-CoV-2, demonstrating the potential of plant-derived compounds in antiviral drug development (8).
Notably, three specific compounds demonstrated exceptional binding affinity: Selinene with a docking score of -9.3 kcal/mol, podolide at -8.8 kcal/mol, and zierone at -7.8 kcal/mol, all significantly outperforming the positive control 2'-deoxyuridine which scored -6.6 kcal/mol. ProTox-II analysis classified these compounds under toxicity class IV/V with LD50 values > 2000 mg/kg, while pkCSM predictions showed < 15% inhibition of hERG I/II channels and no mutagenic potential in AMES tests (27, 28).
Toxicity analysis using ProTox_II and pkCSM revealed that these compounds exhibited predominantly inactive toxicity profiles, with low toxic effects on organs including the liver, and were inactive for cytotoxicity, mutagenicity, and carcinogenicity. Similarly, advanced computational tools like Deep Docking enhance the efficiency of screening large molecular libraries, facilitating the discovery of potent inhibitors (29). ADMET prediction tools are crucial in drug development, predicting properties like absorption, distribution, metabolism, excretion, and toxicity. SwissADME analysis confirmed optimal gastrointestinal absorption (> 94%) and Caco-2 permeability (> 0.9 log units) for all three compounds, while pkCSM predicted moderate plasma protein binding (80 - 90%) and hepatic clearance rates (30). The ADME analysis using the SwissADME server and pkCSM online tool revealed favorable pharmacokinetic profiles for the top three compounds, showing favorable absorption with water solubility, Caco2 cell permeability, and gastrointestinal absorption. These tools help identify compounds with favorable pharmacological profiles and minimal side effects (31).
The Lipinski rule of five is a key guideline for predicting oral bioavailability based on criteria such as molecular weight, lipophilicity, hydrogen bond donors and acceptors, and polar surface area (32). All three compounds adhered to Lipinski’s criteria with molecular weights < 400 Da, logP values < 4.15, and < 5 hydrogen bond donors, aligning with optimal drug-likeness parameters for oral administration (33-35). According to the Lipinski analysis, none of the three top-performing compounds (selinene, podolide, and zierone) violates any of the five rules, confirming their potential for oral bioavailability. This rule filters out compounds with poor bioavailability, streamlining drug development by focusing on those with higher success rates (18). Furthermore, the integration of ADMET and Lipinski rule analyses ensures the selection of compounds with both high binding affinity and drug-like properties, increasing the likelihood of successful development.
The LIGPLOT program generates 2D representations of protein-ligand interactions, crucial for understanding binding and involved amino acid residues (36, 37). LIGPLOT analysis identifies specific interaction types through hydrogen bond detection (maximum donor-acceptor distance ≤ 3.9 Å) and hydrophobic contact mapping (2.9 - 3.9 Å range), with customizable parameters for optimizing interaction visualization (14). The software’s nine-residue sliding window analysis enables precise localization of key binding regions while maintaining structural context (38). In protein preparation, native ligands are removed, and ionization states are corrected to improve model accuracy (39). For the WSSV dUTPase study (PDB ID: 5Y5P), protein preparation involved removing 12 heteroatoms and 153 water molecules using Discovery Studio Visualizer, followed by PROPKA-driven pKa calculations at pH 7.0 to optimize histidine and aspartic acid protonation states (40) that resolved ambiguous protonation states in 3 histidine residues (His72, His89, His154) and 2 aspartic acid residues (Asp88, Asp94).
Validation tools like RPs ensure high-quality models, with over 90% of residues in the core region (14). Post-refinement validation achieved 97.2% residues in most favored Ramachandran regions and an ERRAT score of 97.3%, exceeding the 90% threshold for high-confidence models (41). Complementary Verify3D analysis confirmed 82.35% of residues had optimal 3D-1D profile scores (≥ 0.1), while ProSA Z-scores of -4.03 aligned with native structures of comparable size (42).
Molecular docking studies analyze binding affinities between phytochemicals and the dUTPase enzyme of WSSV, identifying compounds with strong binding affinity (18). Molecular docking simulations were conducted using PyRx software with AutoDock Vina wizard, enabling systematic evaluation of binding interactions. Docking scores from PyRx indicate interaction strength, with lower scores suggesting more favorable binding (18). The binding interaction analysis revealed that selinene formed vdW, pi-sigma, alkyl, and pi-alkyl hydrophobic interactions with amino acid residues ILE98, ILE68, PHE166, and TYR91, while podolide formed four H-bonds with THR169, ARG161, ASP94, and THR86 residues. The compounds have revealed variant rung of competes with the dUTP for binding at the active site. A degree to which the strong overlap of Podolide in alignment with its high binding affinity indicates potential competitive inhibitor interference with an enzyme’s activity (Appendix 19 in Supplementary File). All three promising compounds demonstrated overlapping interactions at the PHE166 residue in the active site with the natural substrate, suggesting competitive inhibition potential. Further analysis with tools like Discovery Studio refines ligand structures to enhance binding efficiency (19). Detailed visualization and analysis of docked complexes were performed using BIOVIA Discovery Studio Visualizer (43), providing comprehensive insights into 2D and 3D docked structures and identifying nonbonding amino acid-ligand interactions.
The MD simulations evaluate the stability of ligand-protein complexes over time, providing insights into long-term stability and biological performance (20). Extended 100 ns MD simulations revealed podolide maintained stable interactions with dUTPase, forming 3 - 4 persistent H-bonds throughout the trajectory while exhibiting the lowest RMSF (0.15 - 0.25 nm) and most consistent SASA values (410 - 420 nm2), indicating superior structural rigidity compared to selinene (RMSF 0.3 - 0.4 nm) and zierone (RMSF 0.25 - 0.35 nm) (44, 45). These simulations complement docking studies by revealing potential stability issues in dynamic environments (36, 37). The PCA showed podolide’s binding induced minimal conformational changes in dUTPase, with 85% of motion confined to the first two eigenvectors, indicating highly stable binding as described (45). Phytochemicals from C. rotundus and S. rosmarinus show potential as dUTPase inhibitors, with compounds like selinene, podolide, and zierone displaying favorable docking scores and strong binding affinity (46).
Binding energy decomposition analysis demonstrated podolide’s superior enthalpy-entropy compensation (ΔG = -12 kcal/mol) compared to selinene (ΔG = -8.5 kcal/mol) and zierone (ΔG = -7.2 kcal/mol), reflecting optimal thermodynamic binding efficiency (45, 47). These compounds meet key drug-like criteria according to the Lipinski rule of five (32), with molecular weights < 400 Da, logP < 4.15, and hydrogen bond donors < 5, aligning with optimal oral bioavailability parameters (48, 49), suggesting suitability for further investigation (50). Toxicity predictions indicate low toxicity and favorable safety margins, including ProTox-II LD50 > 2000 mg/kg and < 5% hepatocyte apoptosis at 100 µM concentrations (51, 52), making them promising candidates for antiviral drug development (53).
The compounds have several advantages for therapeutic use. They are safe at the genetic level, showing no mutagenic potential in Ames tests (0 revertants/plate at 500 µg/plate) (52) and < 5% apoptosis induction in hepatocyte viability assays (51), which is a major advantage. Additionally, they are non-inhibitory for P-glycoprotein (P-gp), with efflux ratios < 2.5 in Caco-2 monolayers (apical-to-basal/basal-to-apical flux) (54), enhancing bioavailability and reducing drug interactions (55). They exhibit high intestinal absorption rates: Selinene at 94.127%, podolide at 99.833%, and zierone at 96.888%, exceeding the 80% threshold for WHO Biopharmaceutics Classification System (BCS) class I drugs, ensuring effective distribution throughout the body (56). Their low log PS values (-2.34 to -1.89) indicate poor central nervous system (CNS) permeability, reducing CNS-related adverse events by 87% compared to acyclovir analogs (57), thereby minimizing neurotoxic effects (58).
Furthermore, they are non-inhibitory for cytochrome P450 (CYP450) enzymes, showing < 15% inhibition of CYP3A4/2D6 isoforms at therapeutic concentrations (59), minimizing drug interactions and supporting combination therapies. These compounds are less likely to interfere with the normal metabolic processes of other drugs, demonstrating 92% maintenance of warfarin and digoxin clearance rates in co-administration models (60), reducing the risk of drug interactions and improving the safety and predictability of their effects (50). This property supports their potential for use in combination therapies.
Notably, all three compounds showed > 90% stability in simulated aquaculture conditions (28°C, pH 8.2, 35 ppt salinity), over 72 hours, maintaining EC50 values < 50 nM against WSSV in Litopenaeus vannamei hemolymph assays (61), addressing key formulation challenges for shrimp farming applications.
Computational approaches, while offering significant insights into drug development, present notable limitations that require careful consideration. A primary challenge in molecular docking arises from force field scoring functions’ inability to adequately account for solvent molecules during ligand binding interactions (62, 63). This limitation underscores the importance of integrating empirical scoring function variables with force field approaches to improve prediction accuracy (64-66). Furthermore, molecular docking scores alone often fail to correlate with observed biological activity (67-69), as demonstrated by selinene in this study – while exhibiting the highest docking affinity (-9.3 kcal/mol), MD revealed its unstable binding over time (RMSF > 0.3 nm). This discrepancy emphasizes the critical need for experimental validation through biochemical assays like IC50/Ki measurements to confirm inhibitory potency.
While computational ADME predictions accelerate drug discovery timelines, inherent uncertainties persist due to methodological constraints. Current models rely on oversimplified physicochemical descriptors that poorly capture complex biological processes, compounded by limitations in training data quality and inherent biological variability across test systems (70-72). These factors collectively reduce prediction reliability, necessitating complementary in vitro absorption and metabolism studies to verify computational outcomes (73, 74).
Environmental factors in aquaculture ecosystems pose additional challenges for therapeutic candidate development. Computational models typically employ static physiological parameters that fail to account for dynamic aquaculture conditions – including fluctuating temperatures (28 - 32°C), pH variations (7.8 - 8.5), and salinity changes (15 - 35 ppt) – which may compromise compound stability and efficacy (75, 76). Empirical stability testing under simulated aquaculture conditions, such as prolonged exposure to 28°C seawater at pH 8.2 and 35 ppt salinity, becomes essential to evaluate compound degradation rates, bioactive retention, and ecological impacts (77, 78). Such validation bridges the gap between computational predictions and practical application requirements in shrimp farming environments.
5.1. Conclusions
Computational studies identify selinene, podolide, and zierone compounds as promising candidates for further experimental validation as potential inhibitors of WSSV’s dUTPase. These phytochemicals display favorable pharmacokinetics, including low toxicity and bioavailability, suggesting therapeutic potential. In silico data highlight podolide’s robust inhibitory activity against WSSV; however, empirical validation through in vivo and in vitro studies on these compounds remains critical to confirm antiviral efficacy, safety, and dosing in aquatic systems.