1. Background
The most important fungal pathogens are yeast species belonging to the genus Candida. Candida species are opportunistic pathogens that can cause superficial mucous membrane infections in life – threatening systemic diseases. Although Candida albicans remains the most frequently cause of Candidiasis, the incidence of the disease caused by non-albicans species has been increased steadily. A major virulence factor of Candida species is its ability to form biofilm. Biofilms are a complex of microorganisms surrounded by a self-production matrix. In fact, biofilms include microbial cells and extra cellular polymeric substances (EPS) (1, 2). Biofilms can organize in most types of surfaces such as rocks and industrial line production; some parts of body like teeth and medical devices (3, 4). They are responsible for a wide range of infections and mortality especially in hospital infections. In the United States more than 45 million medical implant devices are used every year (5). Relevant Infection to these devices occurs in 1-60% of patients. It was reported that Candida species cause 20% of infections. Infections associated with medical devices cause more than 50 % of hospital infections (5). Biofilms exhibit increased resistance against most antimicrobial agents and that is an important problem in treatment of infections related to biofilms (1-6). To treat patients with these kinds of infections, at the first step, it is necessary to identify causes of infection. For this reason, isolation and characterization of yeast from the surface of catheters was carried out.
2. Objectives
The aim of this search was to isolate and identify yeasts from the surfaces of urine catheters of infectious patients in ICU award of Army Family Hospital in Tehran. In addition the ability isolated yeasts to form biofilms was also investigated.
3. Patients and Methods
In this research, 55 urine catheters from patients of ICU award of Army Family Hospital in Tehran were assayed. The catheters were taken aseptically to the laboratory for studying the biofilms. These catheters were sonicated by sonicator (Elma, Germany) in 60 hertz for 2 minutes. Then cell suspensions were cultured on Sabouraud dextrose agar (Merck, Germany) including 0.05 g/L chloram phenicole to eliminate bacterial growth. After that, isolated yeast was cultured on Chrome agar (Himedia), Corn meal agar (Merck). Production of germ tube, assimilation of sugar and PCR method were carried out to identify yeasts at this step. First, yeasts were cultured into sabouraud dextrose broth (Merck, Germany). Next, suspensions of yeasts including 1×10⁶cells/ml were transferred into medium containing (67gr/L) yeast nitrogen base (Himedia, india) medium including 2% agar. Then blank disks were submerged on 20% of sugar solution of glucose, galactose, maltose, lactose, rafinose, terehalose, sucrose, cellobiose and xylose. After that, the disks were placed on the media and production of halo was investigated during 24-48 hours.
For germ tube test, a little of a pure culture of yeast was inoculated to 0.5ml of human serum and incubated for 2 hour. At 10 minutes intervals, a drop of the yeast-serum mixture was placed on a clean microscope slide, covered by a cover slip and observed through 10X and 40X objective lenses of microscope. The appearance of small filaments projecting from the cell surface confirmed formation of germ tubes. The earliest time of each germ tubes production was noted for each serum test (7).
For molecular identification it is necessary to extract DNA of yeasts. Genomic DNA was extracted using the glass bead disruption method. Then PCR was done by a pair of primers ITS1 and ITS4 (Sinagene, Iran). The sequences of the upstream and downstream primers are 5’-TCCGTAGGTGAACCTGCGG 3’ (ITS1) and 5’-TCC TCC GCT TAT TGA TAT GC-3’ (ITS4), respectively.
PCR amplification was carried out in a final volume of 25µl. Each reaction contained 2 µl of template DNA, 12/5µl master mix, 7/5 µl water, 1/5 µl forward primer (ITS1) and 1/5 µl reverse primer (ITS4). An initial denaturation step at 94 °C for 5 min was followed by 34 cycles of denaturation at 94 °C for 45s, annealing at 56 °C for 45s, extension at 72 °C for 55s and final extension step of 72 °C for 10 min. Amplified products were visualized by 1/5 % (W/V) agarose gel electrophoresis in TBE buffer, stained with ethidium bromide and photographed (8, 9).
In this study, the MspI enzyme was able to identify and differentiate six species of all yeasts. The reaction mixture and enzyme digestion was prepared as described in Table 1. All tests were performed at room temperature.
Microtubes were incubated for 2 hour at 37 °C. Restriction fragments were separated on 1/8% agarose gel through electrophoresis for 45 min at 100 V. The gel was stained with ethidium bromide (10).
| Deionized water | 17 |
| X10Buffer | 2 |
| PCR product | 10 |
| MspI enzyme | 1 |
| Total volume | 30 |
Component of the MspI Enzyme Digestion Reaction
4. Results
55 catheters were collected and assayed in this study. 29% of organisms which were isolated from urine catheters surface, were yeasts. 10 out of 16 of isolated yeasts were achieved from female patients (%62.5). It means only 6 yeasts were isolated from male patients (37.5%)
Glucose, galuctose, maltose and terehalose showed the most amount of absorption and were used by all of yeasts. Among these sugars, lactose and rafinose were quantitavely adsorbed less. Results of culturing on Chromagar, Corn meal agar and germ tube technique was summarized in Table 2. As shown in this table, producing of germ tube and presence of chlamydiospore on Corn meal agar was investigated. The color of colony of C. albicans, C. tropicalis, C. krusei, C. glabrata on chrom agar medium were Clear green, blue - Blue-green, pink and purple, respectively.
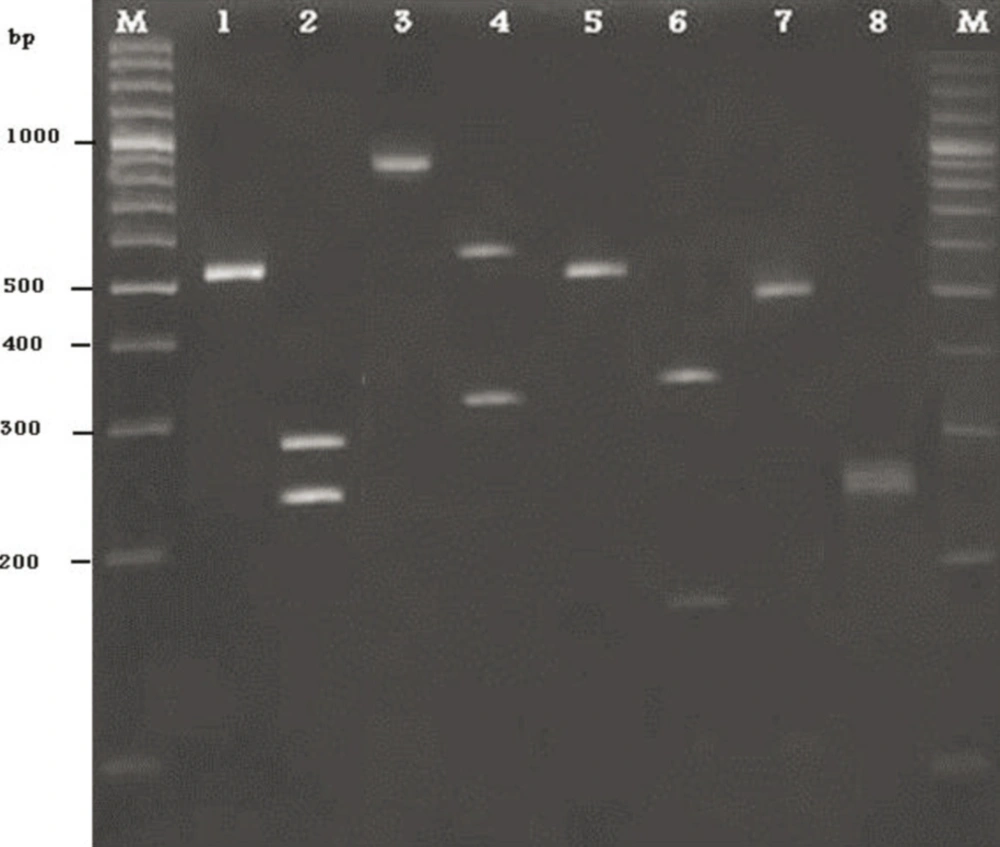
Yeast DNA was amplified by ITS1/ITS4 primers. According to molecular techniques, such as DNA extraction, PCR amplification and sequencing of the intragenic transcribed spacers (ITS) the isolated yeasts were identified as Candida albicans, C. tropicalis, C. krusei and C. glabrata. Among 16 yeasts, 4 out of all isolates were identified as C. albicans and 7, 4, 1 as a C. tropicalis, C. krusei, and C. glabrata, respectively (Figure). In this study, from catheters of patients, 4 different species of Candida were characterized.
| Germ Tube | Corn Meal Agar | |
|---|---|---|
| 1 | + | Producing Chlamydospore |
| 2 | + | Producing Chlamydospore |
| 3 | - | Not producing Chlamydospore |
| 4 | - | Not producing Chlamydospore |
| 5 | - | Not producing Chlamydospore |
| 6 | + | Producing Chlamydospore |
| 7 | - | Not Producing Chlamydospore |
| 8 | - | Not producing Chlamydospore |
| 9 | - | Not producing Chlamydospore |
| 10 | - | Not producing Chlamydospore |
| 11 | - | Not Producing Chlamydospore |
| 12 | + | producing Chlamydospore |
| 13 | - | Not producing Chlamydospore |
| 14 | - | Not producing Chlamydospore |
| 15 | - | Not producing Chlamydospore |
| 16 | - | Not producing Chlamydospore |
Results of Producing Germ Tube, Observing of Chlamydospore on Corn Meal Agar Were Listed in Table.
1) PCR product of C.albicans, 2) PCR product of C.albicans after digestion with the restriction enzyme MspI, 3) PCR product of C.glabrata, 4) PCR product of C.glabrata after digestion with the restriction enzyme MspI, 5) PCR product of C.tropicalis, 6) PCR product of C.tropicalis after digestion with the restriction enzyme MspI, 7) PCR product of C.krusei, 8) PCR product of C.tropicalis after digestion with the restriction enzyme MspI.
5. Discussion
Candida is a fungus that inhabits in half of oral cavities of human (10). A major virulent factor of Candida species is its ability to adhere and form surface attached microbial communities known as biofilms (4, 10, 11). Candida biofilms are more resistant than their planktonic counterparts to different antimicrobial agents (6, 11). Resistance of Candida biofilms to antifungal agents was first demonstrated in 1995 (2). They have crucial role in causing medical devices infections such as stents, shunts, implants and various types of catheters and hospital infections (6, 11). Candida infections of the urinary tract are strongly linked to the presence of urinary catheter (3). In contrast to 13% of non-catheter associated infections, the National Nosocomial Infections Surveillance (NNIS) data indicated that C. albicans caused 21% of catheter associated urinary tract infections (2). Biomaterials infections are a growing alarming problem because they are highly recalcitrant to antimicrobial therapy (11). Recently studies have shown that many of Candida species are able to attach to polymeric surfaces such as urine catheters and produce biofilms.
In this study abundant of isolated yeast species is 25% C. albicans, 25%C. krusei, 6.25% C .glabrata and 43.75% C. tropicalis. Molecular techniques and classical methods to identify isolates are in accordance except of some sugar’s absorption. RFLP–PCR method is a rapid, easy, and reliable technique. This method can also be used in clinical laboratories to identify clinically important Candida spp. (12). Mirhendi and et al. in 2006 used RFLP-PCR for identifying six medically Candida species (13). Ayatollahi Mousavi et al. identified Candida species isolated from oral colonization in Iranian HIV-positive patients by RFLP-PCR method (14). Furthermore, Mohammadi et al. isolated and identified Candida species in patients with various forms of Candidiasis by using this molecular method. Results of all mentioned researches were accordant with our findings (12).
Febre et al. reported that C. albicans with 46.15% frequency was the most abundant fungus followed by C. glabrata (30.77%) and C .krusei (7.7%) from urine specimens (15). These results indicated that C. tropicalis has been replaced with C. albicans and C. tropicalis has the major role to cause yeast infections in form of biofilm. Also in other studies the presence of yeasts in 18.6% of urine specimens of patients with indwelling urinary catheters were observed (15). The results of this study revealed the presence of yeasts in 32% of patients with urinary catheters which indicates the fungal infections ‘increasing. Pakshir was shown in patients with urine catheters involving both bacterial and Candida infections with abundance of 50.4% and 28.7%, respectively (16). Febre et al. have reported, women in comparison to men, are more susceptible to expose yeast infections (15). Pakshir has reported that abundance of Candidiasis in women and men was 68.9 % and 31.1 %, respectively (16). These findings are in accordance with our results. It can be concluded that yeast infections were increased in recent years and the crucial problem of this fungal structure is their resistant to antimicrobial agents because of biofilm formation. Further investigation will be carried out to assay antibiofilm agents against Candida species isolated from surface of urine catheters.