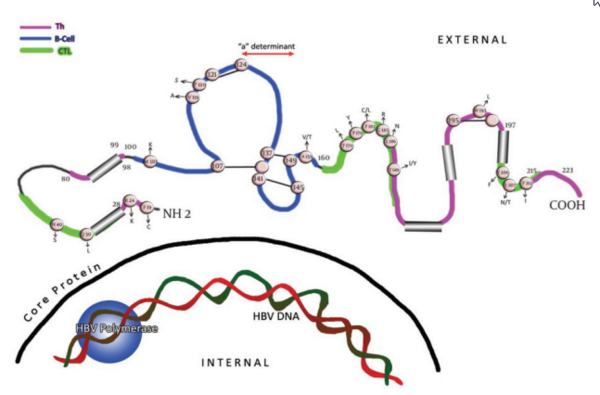
1. Background
Viruses are under constant selection pressure and as a result undergo continual sequence variation. Some of these variants are clinically relevant. The single open reading frame(ORF) that occupies more than one-third of the hepatitis B virus (HBV) genome encodes the three hepatitis B surface antigen (HBsAg)-containing polypeptides. Thus, the three envelope proteins can be translated as; L (large), M (middle), and S (small), or HBsAg. Within the latter, the region between amino acids (aa) 100 and 160 is termed as the major hydrophilic region (MHR). This region is comprised of aa 99-160 that encompass the group-specific “a” determinant (Figure 1). The anti-HBs (antibody to HBsAg) response following natural infection or after immunization is comprised mainly of antibodies that recognize the MHR of the protein. Two major loops and one minor loop are proposed in the MHR, and these are defined by multiple potential disulfide bridges, between aa 107-137 (or 138), 139.147 (or 149), and 121.124 (Figure 1). The majority of the anti-HBs antibodies that appear after a natural infection are directed against this epitope cluster. The antigenic epitopes of the “a" region have been analyzed by binding studies to synthetic peptides using anti-HBs monoclonal/polyclonal antibodies (1-3) and conserved cysteines at positions 124, 137, 139 and 149 (Figure 1) which have been found to be essential for antigenicity and presumably also for the conformation of the protein (4-8). HBV mutations define an obscure phenomenon. In fact, this occurrence seems to be the potential mechanism for the pathogenic basis of chronicity and the clinical complications of this infection. Thanks to the invention of polymerase chain reactions (PCR), facilities for direct sequencing and other molecular approaches, hundreds of reports have been published so far that reveal the relationship between correspondent mutations and the clinical/serological pictures of the chronic patients. Recent studies have shown that HBsAg is more variable than initially thought, and amino acid exchanges are scattered over the whole molecule.
Cysteine residues are shown as white circles and disulphide bridges are indicated as S-S links. The changes selected by antibodies (natural, used in assays and vaccine-induced) that occur upstream and downstream of the determinant are indicated. The carboxyl-terminal end of the protein contains two alpha helices, shown as cylindrical structures.
2. Objectives
The primary aims of this review were to; first, classify different aspects of the HBsAg variants. Second, to describe the patterns of mutational distribution, not only within the “a” determinant in a fashion way, but to present some data on this dispersal outside of that domain, as well as those completely outside of the MHR region. Third, to introduce up-to-date information regarding the clinical and transmission aspects of such variants.
3. Materials and Methods
A comprehensive search of PubMed and ISI was performed with the following Mesh term search keywords: HBsAg mutants and a determinant, vaccine escape HBsAg variants. All published data from 1989 (the discovery of surface mutants) until June 2011, have been included in the study. The inclusion criteria for the study were; mutations affecting vaccine and/or hepatitis B immune globulin (HBIG) escape and horizontal transmission of those variants. Data were analyzed on the basis of amino acid substitutions in different domains of HBsAg in relation to their clinical impact.
4. Results
4.1. HBsAg Escape Mutations
4.1.1. “A” Determinant Mutations
Multiple amino acid changes in the surface-exposed regions of HBsAg, including mutational deletions have been found to abolish the reactivity of monoclonal and polyclonal anti-HBs diagnostic antibodies against the “a” loop epitope cluster, and most anti-HBs in sera from vaccinees are targeted to an epitope between aa 139 and 147 (9).
Amino acid substitution within the MHR can allow replication of HBV in vaccinated persons, as much of the antibodies induced by current vaccines do not recognize critical changes in this surface antigen domain (10). These variants are summarized in Figure 1 and Table 1. In many studies, mutations in vaccinated children were preferentially located in the “a” determinant (11, 12), in contrast to the more randomly located mutations in unvaccinated children (13, 14). In general, replacement in the presumed second “a” determinant loop, including aa positions 144 and 145, are more often associated with anti-HBs immune pressure, than mutations in other epitopic domains of the MHR (Figure 1) (5). On the other hand, second “a” determinant loop mutations are unusual in the absence of anti-HBs pressure (15). Mutations at aa positions 120, 143, 144, and 145 can result in poor reactivity with dly subtyping monoclonal antibodies (16, 17). As this specificity is defined by the residue at aa 122, this indicates the discontinuous nature of many of the epitopes in this antigenic region.
There are linear and discontinuous B-cell epitopes, and some regions are part of more than one epitope. Subtype-related variations can also alter the binding of antibodies to regions linearly distant (16, 17). These differences in the location of mutations may represent the distinctive features of naturally occurring mutations and may suggest the differences in the target epitopes between naturally acquired anti-HBs and those obtained by immunization.
| Amino Acid Position | Wild-Type | Mutant | Cause |
|---|---|---|---|
| 118 | T | A | V a |
| 120 | P | E/S/T/L | V-La-HBIGa |
| 122 | R | L | V |
| 123 | T | N | HBIG |
| 124 | C | R/Y | HBIG |
| 126 | I/T | A/N/S | V-HBIG |
| 127 | P | T/L | V |
| 129 | Q | H/L | V-HBIG |
| 130 | G | D/R | L-HBIG |
| 131 | T | A/N/I/P | V |
| 133 | M | L/I/T | V-HBIG |
| 134 | F/Y | N/R | HBIG |
| 139 | C | Y/S | V-HBIG |
| 141 | K | E/I/R | V |
| 142 | P | S | V |
| 143 | S | L/W | V-HBIG |
| 144 | D | A/E | V-HBIG |
| 145 | G | R/A | V-L-HBIG |
| 146 | N | S | V |
| 147 | C | S | V-HBIG |
| 148 | T | I | V |
| 149 | C | R | V |
| 156 | W | L/C | V-HBIG |
| 157 | A | D/R | V-L |
| 158 | F | Y/S | V-L |
| 179 | F | Y | L |
| 181 | Q | H | V |
| 183 | F | C | V |
| 198 | M | I | L |
| 204 | N | S | V |
| 207 | Q | R/S | L |
| 208 | I | T | V |
| 213 | M | L/I/F | V |
| 218 | I | S/L | V |
Most Frequent Medically Selected Amino Acid Changes in HBsAg.
4.1.2. Mutations outside the “a” Determinant
Changes located outside the “a” determinant region were reported from immunized infants in Singapore born to HBV carrier mothers (18, 19). Interestingly, some of these mutant proteins had reduced binding to monoclonal antibodies against the “a” determinant, and some were most likely transmitted vertically, as they were isolated from both the infant and maternal serum. Their role in vaccination failure is currently unknown. Some of those variants were within the major hydrophilic loop of HBsAg, but mutations at aa positions 183 and 184 were also found. As some of these mutations had decreased binding to an “a”-specific monoclonal antibody, their functional analysis will contribute to the understanding of the antigenic structure of the HBV envelope. Likewise, multiple aa changes in the surface-exposed regions of HBsAg, including mutations and/or deletions upstream and downstream of the “a” determinant region (Figure 1) (20-22), were found to abolish the reactivity of monoclonal and polyclonal anti-HBs diagnostic antibodies against the “a” loop epitope cluster, and they were not recognized by the vaccinee’s sera either (2, 3, 26). The region between aa 118 and 123 was identified as a hot spot for insertions by investigators, illustrating its immunological importance (2, 24, 25). Furthermore, aa substitutions have been found in residues downstream (38 to 98) and upstream (between 164-215) of the MHR region of native and or recombinant surface proteins with different binding capacity to the antibodies (27-31).
In an attempt to study the sensitivity of modern assays for the detection of HBsAg, investigators have evaluated the performance of different assays for the detection of single, double and triple recombinant and/or native HBsAg mutants (28, 30-34). Assays were challenged with native and/or recombinant HBsAg in positions upstream and downstream of the “a” determinant (Figure 1). Interestingly, the majority of HBsAg tests were able to detect the mutants (data are not shown).
4.1.3. Mutations in Surface Protein T cell Epitopes
Being a structural protein, HBsAg is an immune target. The distribution of the mutations within known surface protein immune epitopes reflects the virus-host interaction over a prolonged infection period. HBV surface proteins contain both B and T cell antigenic epitopes. Mutations in T cell epitopes may theoretically influence the anti-HBs antibody profile through an interaction between CD4+ helper T cells and B cells (5). Appropriate reactivity of T cells is a prerequisite for adequate anti-HBs production after infection with the HBV, as well as after hepatitis B vaccination. Thus, the T cell epitopes of HBsAg, being targets for recognition by T cells, should also be affected. The humoral response to HBsAg is T cell-dependent. At least four regions within the HBsAg present epitopes for MHC class II restricted CD4+ T cells (Table 2) (35, 36).
The effect of HBsAg T cell epitope variants on the cellular immune response has been studied in individuals vaccinated against HBV by Bauer et al. (37). Six of the 23 different variants in two HBsAg Th epitopes were shown to be responsible for inadequate T cell reactivity. Similarly, the MHC class I-restricted CDB+CTL response plays a key role in suppressing HBV infections. Schirmbeck et al. (38) showed that in H-2b mice even small changes in amino acid residues within the two different CTL epitopes that mimic natural variants of adw2, ayr and adr, completely eliminated the immunogenicity of each epitope. A majority of mutations found up- and downstream of MHR in different studies were found to be potentially responsible for vaccine breakthroughs. HBsAg undetectability (2, 3, 20, 23, 39) were found to be located within the known, either Th or CTL (or both), epitopes of the surface protein (Table 2) (35, 36, 40, 41).
In general, viral mutations in CTL epitopes were able to evade cellular immunity and thus contribute to persistency (42, 43). Naturally occurring mutations within the CTL epitope could lead to epitope inactivation and T cell receptor antagonism (44). Studies from our laboratory have revealed a high frequency of mutations at aa positions of the surface gene that coincides with the HLA-restricted CTL epitope in patients with chronic hepatitis B, suggesting that these mutations might contribute to chronic infections (45). Nonetheless, accumulating evidence indicates that the genomic heterogeneity of the virus does not account for HBsAg negative status in all cases (25, 39, 46).
| 100-160 | B | non HLA restriction | (91) |
| 19-28 | Th | Class Π | (34) |
| 28-51 | CTL | Class I HLA-A2 | (40) |
| 80-98 | Th | Class Π | (34) |
| 171-179 | CTL | Class І | (39) |
| 175-184 | CTL | Class I HLA-A2 | (35) |
| 186-197 | Th | Class Π | (35) |
| 206-215 | CTL | Class І HLA-A2 | (35) |
| 215-223 | Th | Class Π | (35) |
Proposed Antigenic Epitopes within HBsAg.
4.1.4. Mutation in Other HBV Proteins
In other studies, however, the failure of HBsAg detection was not fully explained by surface (S) gene mutations (47, 48). Furthermore, multiple mutations outside the surface protein could be found within the core, pre-S, X and polymerase with known functional and/or immune epitope reactivity (39, 49, 50).
Multiple alterations in the genome possibly have a synergistic effect in the downregulation of HBsAg production, and an isolated cause for mutation in a particular gene or regulatory region could not be established. Thus, it appears that the nondetectability of HBsAg in serum may arise as a result of several mechanisms caused by alterations in the; structural, functional, and regulatory regions of the HBV genome, causing it to lie below the sensitivity of standard enzyme-linked immunosorbent assay tests (46, 51, 52). Mutations outside the surface protein may also influence HBV replication capacity. According to earlier studies, these mutants have also been reported to be less ‘‘replication fit,’’ compared to wild-type viruses in vitro, providing a plausible explanation for the low HBV DNA levels (53, 54).
5. Clinical Aspects of HBsAg Mutants
5.1. Chronic State
Little is known about the natural history of HBsAg variants. The presence or absence of HBsAg variants in chronic patients (i.e., children, inactive carriers, chronic active carriers, etc.) has created a confounding scenario. Some reports have suggested that patients with pure or mixed “a” epitope variants had lower alanine transaminase (ALT) levels compared with those with wild-type HBV-DNA (55). Asian–Indian patients with HBsAg variants had a prolonged illness in the form of anicteric chronic hepatitis. These patients had a higher frequency of quiescent cirrhosis and hepatocellular carcinoma than those with the wild-type hepatitis B virus. On the other hand, S-gene mutants have been associated with an aggressive or worsening clinical course in some reports (56, 57). Further analysis is required to assess the association of “a” epitope variants with the development of liver diseases. A number of escape-variant viruses during the course of chronic infection within the “a” determinant were reported to be replicated in spite of the presence of anti-HBs (15, 39, 58-68). This phenomenon, which has been explored by molecular approaches in vaccinated individuals, may result from at least seven reasons. First, individuals protected by HBV vaccines had a low capacity of immune response (in early childhood) and they could not develop enough anti-HBs antibodies. Second, the blood of newborn babies had already been contaminated with HBV particles in the uterus, so that the HBV vaccines were ineffective. The third situation could be the infection of HBV mutants or simultaneous infections with wild types. These mutants may present as a small percentage, a minor strain together with the predominant wild-type strain, either before the onset of antibody-mediated immune pressure and thus are merely selected, or they may emerge under conditions unfavorable for wild-type virus, which cannot be detected through the use of sequencing analysis. Fourth, mutations within the surface gene are one of the factors contributing to a loss of HBsAg detection by immunoassay (diagnostic-escape mutants). These “a” determinant variants may go undetected by conventional HBsAg screening tests. Sixth, the anti-HBs in these patients may possibly be directed against the epitopes outside the “a” determinant, such as d/y or w/r subtype determinants (66). Seventh, low expression of HBsAg due to a low viral load (which might be just enough for viral assembly, but underneath the sensitivity of standard tests) thus moving it below the sensitivity of standard serologic assays (69). In some cases, mutations in the “a” determinant have been seen without the presence of anti-HBs. The lack of serum anti-HBs in these patients does not necessarily indicate a lack of humoral immune responses to the “a” determinant, because it has been reported that anti-HBs can be complexed with HBsAg (and detection of one or the other alone merely indicates dominance at that time) (66, 70, 71). Therefore, the data suggests that escape mutations could be induced by immunological pressure exerted on the MHR (within or up and downstream of the “a” determinant), even in patients who are found to be negative for serum anti-HBs through the use of the usual assay.
5.2. Transmission
Blood safety issues related to the safety of the blood or tissues of an HBsAg-negative donor and the risk of transmission are of grave concern. There is a fear that escape mutants may be spread by blood transfusion, since HBV DNA detection by nucleic acid amplification technology (NAT) is not mandatory for blood donor screening in many countries, especially in those areas where the prevalence of escape mutants is expected to be high. Earlier studies clearly indicated that the transfusion of blood containing HBsAg has been associated with the development of post transfusion hepatitis B (72-74), and that cases of HBV infection have been identified after transfusion of a HBsAg negative blood supply (75-77). Moreover, it is well known that mutants of HBsAg are able to cause infection and horizontal transmission despite the presence of anti-HBs (78-82).
Studies in both animals and humans have proven that the G145R mutant can infect the host without the help of a wild type virus (83, 84). This suggests that the native conformation of the second ‘‘a’’ loop is not essential, at least for the entrance of the virus into the cell. As a consequence it may be argued that the second ‘‘a’’-loop related antibodies may not be protective on their own, although they may lower the viral load in the circulation by the formation of aggregates that are easily opsonized (81). Escape mutants may breakthrough vaccine programs and spread locally (85). The issue of whether the current vaccine is effective in preventing infection by these mutants, particularly in infants born to HBV carrier mothers, is currently under debate (13, 42, 86, 87). Of particular concern is the observed emergence of “a” determinant mutants occurring in geographic regions where universal vaccination has been instituted (14, 88).
Although the absolute number of new infections has been reduced in these regions, infections are most often observed in vaccinated infants born to HBV carrier mothers, suggesting that infants exposed to a maternal mutant virus may select for this strain during the development of vaccine-induced antibodies (39, 89). An epidemiological model of the HBV that investigated possible patterns of emergence of a vaccine-resistant strain, showed that using pessimistic assumptions (i.e., the current vaccine provides no cross-immunity against the variant), the variant would not become dominant over the wild-type for at least 50 years (90).
6. Conclusions
In conclusion, mutations have been described in all of the four open reading frames of the HBV. Thus, from a clinical perspective, the S escape mutant is the most troublesome, because in the absence of surveillance systems and/or a high index of suspicion, a diagnosis can be difficult to establish. A HBV genome containing mutated immune epitopes can no longer be recognized by specific T cells of the host immune surveillance and they will not enhance anti-HBs production; this could lead to the progression of chronicity of the HBV infection.