1. Background
Pseudomonas aeruginosa is a non-fermenting aerobic gram negative microorganism that is identified in clinical specimens of hospitalized patients and their environments. According to the data released by Center for Disease Control (USA), P. aeruginosa is the fifth most common pathogen among hospital microorganisms which causes 10% of all hospital acquired infections (HAI) (1-3). This organism has a considerable compatibility ability with the environment and acquires antibiotic resistant genes and can be transmitted in hospital environment from one patient to another (4, 5). Because of an increase in the antibiotic resistance in these bacteria, especially multi drugs ones, many problems have been created after the treatment of infections by the pathogens (6).
The epidemiologic study showed that the infection caused by P. aeruginosa can lead to such drawbacks as a need for a surgery, an increase in the length of hospitalization, a continuous supervision, an increase in treatment expenses and finally increased mortality rates (7). Resistance mechanisms in these bacteria are explainable by both the mutation of genes and the change of action mechanisms in chromosomal and transferable gene elements like transposons or integrons (8). Integrons are transportable genetic particles which can carry the antibiotic resistance genes (9). These particles are located in many different parts of plasmids and chromosomes.
Integrons can also surround genes and transfer them while they are in the gene cassettes (10). Integrons include integrase gene, two stable conservative reigns called sul1 and int1 as well as one variable region of gene cassettes. Integrons are classified into four classes according to their integrase genes. The prevalence of class 1 integron is high and has been studied more than the other classes (11). Because integrons are located on plasmids, they can easily spread among bacteria species rapidly. The resistant genes which are located on gene cassettes can be separated and penetrated into other integrons. This is a notable phenomenon in the creation and distribution of new resistant cassettes and the evolution of plasmids and chromosomes.
The importance of the existence of multi-antibiotic resistance and the integrons play important roles in multiple resistances (12, 13). While new antibiotics are available, but the appearance of multidrug resistant bacteria, especially in nosocomial infections, can cause many problems around the world, so the consciousness of local patterns of antimicrobial resistance for the treatment of these infections is necessary due to the fact that their distribution and antibiotic resistance vary in different times and among different areas.
2. Objectives
The aim of the present study was to find out the molecular relation, the existence of integrons with sensitivity pattern and multidrug resistance of P. aeruginosa strains isolated from the environment and the equipment in ICU, at Babol Medical University in north of Iran.
3. Materials and Methods
3.1. Bacterial Isolates
This cross-sectional study was performed on 33 P. aeruginosa isolates, collected from different parts of environment and equipment in ICU at Shahid Beheshti Hospital affiliated to Babol Medical University in north of Iran from 2008 to 2009. At first, the humid soap (mixed with sterile distilled water or nutrient broth media) was pulled completely on the considered surface and after putting it in transport medium, it was sent to the microbiology laboratory. Samples were cultured on nutrient agar (Merck, Germany) immediately and then, the media were incubated at 37 °C for 18 to 24 hours. Then, the purred colonies from each sample were identified as P. aeruginosa with differential tests, collected in BHI broth (Heart Infusion Broth, Difco) and two 1.5 ml micro tubes; in which one of them contained 1 mL distilled water and the other one 1 mL physiological serum. These samples were kept in -20 ºC for performing sensitivity test and DNA extraction.
3.2. Antimicrobial Susceptibility Testing:
Antimicrobial susceptibility testing was performed by disk agar diffusion method (DD) according to the standard protocol of CLSI 2006 M2-A9 recommendation (14). The following antimicrobial disks (HiMedia, India) were used for antimicrobial susceptibility testing; cefepime (FEP: 30 µg), ceftriaxone (CRO:30 μg), cefazolin (CZ: 30 μg), ceftazidime (CAZ:30 μg), ceftizoxime (CT: 30 μg), imipenem (IPM: 10 µg), ticarcillin (TIC: 75 μg), cefotaxime (CTX:30 μg), amikacin (AN:30 µg), ofloxacin (OFX: 5 μg), ciprofloxacin (CRO: 30 µg) and gentamicin (GM: 10 µg).
3.3. Determination of Minimum Inhibitory Concentration
Minimum Inhibitory Concentrations (MICs) were determined by micro broth dilution method according to the recommendations of the standard protocol of CLSI 2006 M07- A7 (15).The antibiotics were purchased from Sigma Chemical Company.
3.4. DNA Extraction
For the extraction of DNA, the high pure PCR template preparation kit from the Roche, the German company, was used. After bringing out the DNA, each sample was kept in -20 °C until the PCR stage.
3.5. Class 1 Integron PCR
The primer sequences designed, were R5'-AGG AGA TCC GAA GAC CTC-3', F5'-TCT CGG GTA ACA TCA AGG-3' for the amplification of intl gene, which could produce 234 bases in per nucleotide length segment. The PCR reaction was performed in 50 μL volumes which contained 10 µL extractions of DNA (equal to 1 μg), 5 pmol/L from each primer, 1.5 mmol/L MgCl2, 0.2 mmol/L dNTPs and 1.5 unit of Taq DNA polymerase enzyme (13).The amplification reactions for int1 gene, included primary denaturation at 94 ºC for five minutes and then, was followed by 35 cycles of denaturation at 94 ºC for one minute, annealing at 53 ºC for one minute and extending at 72 ºC for 30 seconds. Moreover one cycle for the final extension at 72 ºC for five minutes was performed (13).
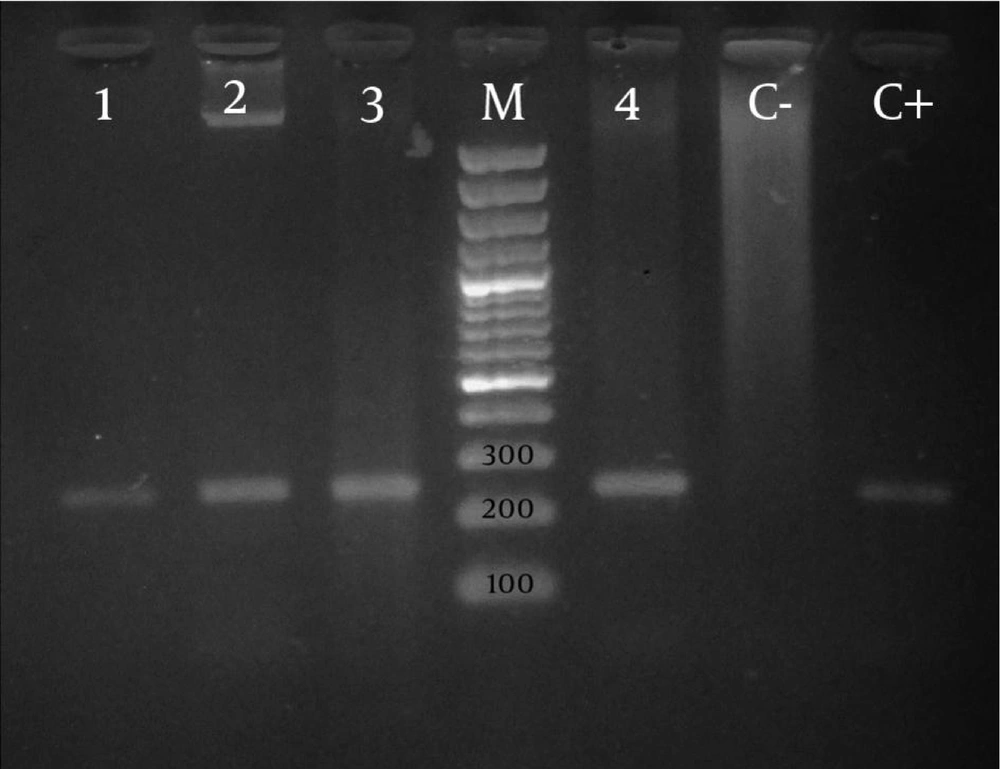
PCR reaction was conducted in the presence of positive and negative controls. After performing PCR reaction, electrophoresis of PCR products was conducted in 1.5% agarose gel for 60 minutes. Then, the results were evaluated under UV light on the UV gel document. After PCR, the specific band with associate weight was considered as a fragment of intl gene.
3.6. Statistical Analysis
The data was analyzed using the SPSS statistical software version 18. The Chi-square test was employed to calculate the P value in terms of resistant, intermediate, and susceptible numbers of integron-positive and negative isolates. The χ2 test, or the Fishers’ exact test, when appropriate, was used in a univariate analysis to assess the differences between two groups of isolates. P values less than 0.05 were considered as statistically significant.
4. Results
Thirteen of 33 (39.4%) of P. aeruginosa species had intl genes among which 24.2% were multidrug-resistant and 15.2% were intermediate or sensitive. The existence of the gene and the resistance to these antibiotics were examined by DD and MIC methods, which were shown in Table 1 and Table 2 , respectively. No significant differences were seen between the presence of integron gene and the resistance to the antibiotics except ofloxacin. The PCR result with the mentioned primer is one segment with 243 base pairs ( Figure ).
Most resistance was found against cefepime 100% and ceftazidime 97% and the lowest was against ciprofloxacin and ofloxacin 42.5% ( Table 2 ). Most of these strains (97%) were resistant to more than four antibiotics. To compare disk diffusion and micro dilution methods, the most likely change of resistance against antibiotics has been found for ceftazidime with 97% compared to 69.6% which was statistically significant, P =0/001 ( Table 1 and Table 2 ).
| DD (n = 33) | Integron Positive (n = 13) | Integron Negative (n = 20) | P value a | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S% (n) | I%( n) | R% (n) | S% (n) | I% (n) | R% (n) | S% (n) | I% (n) | R% (n) | ||
| IPM | 45.5 (15) | 3 (1) | 51.5 (17) | 23.1 (3) | 0 (0) | 76.9 (10) | 60 (12) | 5 (1) | 35 (7) | 0.02 |
| FEP | 0 (0) | 0 (0) | 100 (33) | 0 (0) | 0 (0) | 100 (33) | 0 (0) | 0 (0) | 100 (20) | NS |
| TIC | 48.5 (16) | 0 (0) | 51.5 (17) | 30.8 (4) | 0 (0) | 69.2 (9) | 60 (12) | 0 (0) | 40 (8) | NS |
| OFX | 51.5 (17) | 6 (2) | 42.5 (14) | 23.1 (3) | 0 (0) | 76.9 (10) | 70 (14) | 10 (2) | 20 (4) | 0.001 |
| AN | 45.5 (15) | 0 (0) | 54.5 (18) | 30.8 (4) | 0 (0) | 69.2 (9) | 55 (11) | 0 (0) | 45 (9) | NS |
| GM | 36.5 (12) | 9 (3) | 54.5 (18) | 15.4 (2) | 7.7 (1) | 76.9 (10) | 50 (10) | 10 (2) | 40(8) | 0.02 |
| CZ | 6 (2) | 0 (0) | 94 (31) | 0 (0) | 0 (0) | 100 (13) | 10 (2) | 0 (0) | 90 (18) | NS |
| CRO | 21.2 (7) | 21.3 (7) | 57.5 (15) | 15.4 (2) | 7.7 (1) | 79.9 (10) | 25 (5) | 30 (6) | 55(11) | 0.07 |
| CT | 27.3 (9) | 0 (0) | 72.7 (24) | 23.1 (3) | 0 (0) | 76.9 (10) | 30 (6) | 0 (0) | 70 (14) | NS |
| CTX | 27.3 (9) | 12.1 (4) | 60.6 (20) | 23.1 (3) | 0 (0) | 76.9 (10) | 30 (6) | 20 (4) | 50 (10) | NS |
| CIP | 48.5 (16) | 9 (3) | 42.5 (14) | 30.8 (4) | 7.7 (1) | 61.6 (8) | 60 (12) | 10 (2) | 30 (6) | 0.07 |
| CAZ | 3 (1) | 0 (0) | 97 (32) | 0 (0) | 0 (0) | 100 (13) | 5 (1) | 0 (0) | 95 (19) | NS |
a P-value was calculated using the Chi square test in terms of the number of resistant strains and susceptible strains in the integron-positive and integron-negative groups.
| DD (n = 33) | Integron Positive (n = 13) | Integron Negative(n = 20) | P value a | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S% (n) | I% (n) | R% (n) | S% (n) | I% (n) | R% (n) | S% (n) | I% (n) | R% (n) | ||
| IPM | 48.5 (16) | 15.2 (5) | 36.3 (12) | 38.5 (5) | 23.1 (3) | 38.5 (5) | 55 (11) | 10 (2) | 35 (7) | NS |
| FEP | 6 (2) | 3 (1) | 91 (30) | 0 (0) | 0 (0) | 100 (13) | 10 (2) | 5 (1) | 85 (17) | NS |
| TIC | 51.5 (17) | 3 (1) | 45.5 (15) | 38.5 (5) | 0 (0) | 61.5 (8) | 60 (12) | 5 (1) | 35 (7) | NS |
| OFX | 48.5 (16) | 0 (0) | 51.5 (17) | 23.1 (3) | 0 (0) | 76.9 (10) | 65 (13) | 0 (0) | 35 (7) | 0.001 |
| AN | 42.4 (14) | 21.3 (7) | 36.3 (12) | 30.8 (4) | 23.1 (3) | 46.1 (6) | 50 (10) | 20 (4) | 30 (6) | NS |
| GM | 36.3 (12) | 3 (1) | 60.7 (20) | 15.4 (2) | 7.7 (1) | 76.9 (10) | 52.6 (10) | 0 (0) | 52.6 (10) | 0.065 |
| CZ | 12.1 (4) | 0 (0) | 87.9 (29) | 7.7 (1) | 0 (0) | 92.3 (12) | 15 (3) | 0 (0) | 85 (17) | NS |
| CRO | 30.3 (10) | 0 (0) | 69.7 (23) | 23.1 (3) | 0 (0) | 76.9 (10) | 35 (7) | 0 (0) | 65 (13) | NS |
| CT | 27.3 (9) | 0 (0) | 72.7 (24) | 23.1 (3) | 0 (0) | 76.9 (10) | 30 (6) | 0 (0) | 70 (14) | NS |
| CTX | 6 (2) | 21.3 (7) | 72.7 (24) | 0 (0) | 15.4 (2) | 84.6 (11) | 10 (2) | 25 (5) | 65 (13) | NS |
| CIP | 54.5 (18) | 0 (0) | 45.5 (15) | 38.5 (5) | 0 (0) | 61.5 (8) | 65 (13) | 0 (0) | 35 (7) | NS |
| CAZ | 18 (6) | 12.3 (4) | 69.7 (23) | 0 (0) | 7.7 (1) | 100 (13) | 30 (6) | 15 (3) | 55 (11) | 0.018 |
aP-value was calculated using the Chi square test in terms of the number of resistant strains and susceptible strains in the integron-positive and integron-negative groups.
5. Discussion
Whereas, integron class 1 plays an important role in creating and transferring the antibiotics resistance, its prevalence is an alarm for infections caused by this bacterium. The present study investigating the existence of integron revealed that 39.4% of the isolates contained integron classes 1. This is comparable with previously reported frequencies of 40.8% in China, 41.5% in Brazil (P. aeruginosa), 60% in the United Kingdom (Acinetobacter baumannii), 43% in Europe (Gram-negative isolates), 50% in The Netherlands (Enterobacteriaceae), 59% in France (Enterobacteriaceae), and 52% in Taiwan (Escherichia coli) (16, 17).
The reported percentages were higher compared to our study, which can be all due to the differences among geographical regions and the bacteria strains. The antibiotic resistance rates in class 1 integron-positive strains of P. aeruginosa were noticeably higher than those in class 1 integron-negative strains, which were in concordance with another study performed in China (11). While all isolated P. aeruginosa, resistant to cefepime, have had class 1 integron gene, in this study, the integrons were significantly associated with resistance to ofloxacin ( Table 1 and Table 2 ).
In another study, there was a significant relationship between resistance and some antibiotics like aminoglycosides, beta–lactam and quinolons agents (16, 18). This is not surprising, since many antibiotic resistance gene cassettes encoding resistance to a wide range of antibiotics have been reported previously. Thus, the intensification of resistance in isolated strains in different places showed the expansion of integrons in gram negative bacteria (11, 13, 19-21). Furthermore, the presence of integrons among different classes of antibiotics can cause serious problems in the distribution of this resistance.
Despite the fact, the class 1 integron has not been seen in some strains of P. aeruginosa, which is resistant to antibiotics. In fact, it is well known that multiple mechanisms, such as chromosome mutations, acquisition from plasmids or existence of integrons except class 1 are related to antibiotic resistance in this species of P. aeruginosa. The results of our study showed that 100% of Pseudomonas strains isolated were resistant to cefepime and 97% against ceftazidime ( Table 1 ).
In some studies performed in Tehran, Iran, 100% of strains were resistant to cefepime (22). Resistance rate to ceftazidime in Europe ranges from 15% to 97% and to imipenem less than 1% up to 85% (23). In Saudi Arabia, the susceptibility of P. aeruginosa, significantly declined after 2007, especially for carbapenem (66% in 2004 to 26% in 2009), ceftazidime (69% in 2004 to 44% in 2009), and ciprofloxacin (67% to 49%) (24). In other antibiotics, the resistance rate was different in different places of the world. The reported resistance against ceftazidime was 9% in France, 12.3% in Brazil, 26% in Turkey, 4.6% in Japan, 35% in Russia, 12% in Canada, 11.1% in United States and 15% in Spain (25).
A review of the 1999 National Nosocomial Infection Surveillance (NNIS) survey data from the CDC showed that imipenem resistance among P. aeruginosa strains had increased from 12% to 19% and quinolone resistance in P. aeruginosa from 12% to 23% (24). Furthermore, the antibiotic susceptibility test in P. aeruginosa showed that 97% of isolated strains were resistant to more than four antibiotics and among them, 36.4% were resistant to all tested 12 antibiotics. Multidrug resistance mechanism in P. aeruginosa is complicated and can cause many changes in permeable membranes, plasmids, integrons and so on (9). Although all strains of multi-drug resistance of P. aeruginosa were isolated from ICU equipment and the hospital environment, many factors such as lack of proper hygiene in hospital environment and equipment, inappropriate information of workers at the Hygiene Service Department and the way of controlling the infection can transfer the antibiotic resistant among bacteria’s colonies, but integrons can transfer the resistant gene among bacteria by new distribution mechanisms (26).
Our results showed a high prevalence of class 1 integron gene in most of P. aeruginosa strains isolated from different parts of the environment and equipment in ICU. The role of these transferable genetic agents was proven in the creation of resistance. So, the environmental bacteria represent a reservoir for the dissemination of clinically relevant multidrug-resistant antibiotics and should be taken under control to reduce the appearance or distribution of these antibiotic resistant agents.