1. Background
Chloroplasts are major sources of food productivity and life-sustaining for whole organisms on the planet earth. Plastids are known as semi-autonomous organelles with a small, 120 kb to 160 kb genome which exists in higher plant cells with about 1000 to10000 copies per cell (1, 2). They have their own transcription and translation machinery (2-4). The chloroplast genome (plastome) usually consists of two copies of inverted repeats (IRA and IRB), a large and a small single copy region (LSC and SSC, respectively) (3, 5).
Insertion of transgene into the chloroplast genome has become an attractive alternative to nuclear transformation. Several advantages such as accumulation of high levels of foreign proteins, the feasibility of expressing multiple proteins from polycistronic mRNAs, gene containment through the lack of plastids in mature pollens, biosafety and absence of epigenetic transgene instability effects are some of the chloroplast engineering profits. Considering these advantages, induction of transplastomic plants could be useful for conferring desirable agronomic traits, metabolic engineering and producing biopharmaceuticals (6-8). Chloroplast transformation was generally accomplished by the biolistic method, with the E. coli vectors which contains a selectable marker gene, homologous recombination regions and the gene of interest (1, 8, 9). This technology is established and can be routinely used in tobacco transformation; But now, successful plastid transformation was reported in other plants such as rice (Oryza sativa), soybean (Glycine max), eggplant (Solanum melongena L.), cauliflower (Brassica oleracea), cabbage (Brassica capitata), lettuce (Lactuca sativa), oilseed rape (Brassica napus), petunia (Petunia hybrid), poplar (Populus spp.), potato (Solanum tuberosum), tomato (Solanum lycopersicum), carrot (Daucus carota) and cotton (Gossypium hirsutum) (8, 10, 11).
Today some plant species are being used as factories for producing valuable biomaterials and essential industrial enzymes and proteins. Regarding the high capacity of chloroplasts to express and also accumulate foreign protein, researchers are interested in using transplastomic plants for production of edible vaccines, antibodies (plantibodies), and therapeutic substances (12, 13). Spinach is an important edible vegetable which can be cultivated under short day photoperiods and it has become as a plant model for chloroplast genetic engineering investigations. Nuclear transformation in spinach has been reported by several researchers (14-16). Spinach leaves are large with a dark green color which indicates high number of chloroplasts per cell. Spinach plastome is a double-stranded DNA molecule with 150725 base pairs (17). Number and size of chloroplasts are different, and its appearance depends on genotype characteristics. Generally a single mesophyll cell has fifty to more than a hundred chloroplasts (17). Apical meristem cells in spinach have about twelve proplastids. When cell expansion occurs, chloroplast starts dividing which forces the leaf growth promotion. The number of chloroplasts per cell reaches about 200 and each individual chloroplast consists of 57 to 353 plastome.(18). So transgene integration into spinach chloroplast has been anticipated to yield a high expression of gene of interest.
The first step of chloroplast transformation is to design and construct specific vectors. Transgene can be integrated into the plastome by homologous recombination between flanking sequences on vector and analogous sequences on plastome. In this research, the attempt was to select, isolate and clone these flanking sequences from spinach plastome to construct spinach chloroplast vector. Chloroplast structure is similar to the bacteria due to its prokaryotic origin. This suggests that the transcription and translation system of bacteria can recognize the chloroplastic regulatory elements. It is believed that such phenomenon is attributed to endosymbiosis contemplate evolutionary. Thus, by knowing such concept the attempt was to test the constructed vector in E. coli. The most common selectable marker for chloroplast transformation is the aadA gene. It encodes aminoglycoside 3΄-adenylyltransferase which is responsible for both spectinomycin and streptomycin resistances. This marker is generally used to construct the chloroplast vector (8, 15, 19). Also 16S rRNA promoter (Prrn) and rbcL 3΄UTR are putative promoter and terminator in chloroplasts (9).
2. Objectives
In this study, an investigational report of bacterial aadA gene which is controlled by the tobacco chloroplast Prrn promoter and the Chlamydomonas rbcL 3΄UTR terminator was presented that can successfully express aadA gene in E. coli.
3. Materials and Methods
3.1. Bacteria
Escherichia coli strain DH5α was employed as a host for cloning and testing the construct.
3.2. DNA Extraction
Spinach seeds were planted in selected pots under greenhouse condition. All requirements for plant growth regarding food, light, water and other factors were provided according to plant needs. When plant seedlings were reached to an appropriate stage, fresh leaves were harvested to extract total DNA. Approximately 0.5 gram of fresh leaves was powdered in oven containing liquid nitrogen using a mortar and pestle. Then 900 µl of extraction buffer was added and transferred to 1.5 ml microtube and inverted several times. 100 µl of sarcosine 10% (w/v) was added to microtube and kept in 65°C for half an hour and then centrifuged in 13000 rpm for 10 minutes. 800 µl of the supernatant was transferred to a new microtube and 100 µl of 5M NaCl plus 200 µl of CTAB (cetyl trimethyl ammonium bromide)5% (w/v) were added. The microtube kept in 65°C for 10 minutes. Then the chloroform : isoamyl alcohol solution (24:1) was added in the equal volume of microtube solution and mixed gently and centrifuged in 13000 rpm for 10 minutes, this step repeated once again. The supernatant was transferred to a new microtube and the equal volume of isopropanol added and kept in -80°C for 15 minutes and then centrifuged in 13000 rpm for 10 minutes. The supernatant was removed and the DNA precipitated with 100 µl ethanol by centrifugation in 13000 rpm for 10 minutes. Then the supernatant was removed and 50 µl of dioneized water added to the extracted DNA after ethanol evaporation and drying. The extracted DNA was kept in -20°C.
3.3. Primer Designing
Two specific primers were designed to isolate flanking sequences, containing the intergenic region of spinach plastome, between the trnI and the trnA genes (nucleotides 99040-102040; GenBank accession number AJ400848). The total size of these flanking regions is about 3 kb. Designed primers were PLF (5 ́GCGGCCGCAACAGACTGCCGGTGATAAG 3 ́) and PRF (5 ́ GGCGCGCCTCAGAGCATAAGCTAGTG 3 ́). NotI and AscI restriction sites were used in 5΄ end of PLF and PRF sequence respectively. aadA expression cassette was amplified by two specific primers (FAADA: 5΄ CCGCGGTTAATTAACAGACTAGTGGAT CGCAC 3΄ and RAADA: 5΄ GGTACCGTTTAAACCATTGCTAGCGGCAATTC 3΄). SacII, PacI and KpnI, PmeI restriction sites were used added in 5΄ end of FAADA and RAADA respectively.
3.4. Vector
pUC19 (Invitrogen, USA) was used as a backbone vector in this investigation.
3.5. Cloning of the Fragments in pUC19
The 5 ́ end of primers was phosphorylated via T4 Polynucleotide Kinase (Fermentas, Lithuania) before using PCR. The INSL and INSR sequences were amplified by high fidelity PCR Enzyme Mix (Fermentas, Lithuania) using PLF and PRF primers and total DNA of spinach as a template. PCR products were extracted using PCR purification kit (Bioneer, South Korea). The end of PCR products were filled by Klenow Fragment (Fermentas, Lithuania). pUC19 vector was digested with PvuII (Fermentas, Lithuania) and was purified with gel extraction kit (QIAGEN, Germany). The digested vector was dephosphorylated with shrimp alkaline phosphatase (Fermentas, Lithuania) to avoid self-ligation. Finally, the PCR product was blunt-end cloned into the PvuIIsite of pUC19, forming the pUCINS (pUC+INSL+INSR).
The selectable marker expression cassette (aadA gene plus ribosome-binding site, Prrn promoter and rbcL 3΄UTR) was amplified by PCR using FAADA and RAADA primers. The end of PCR product was filled by Klenow Fragment. The pUCINS digested with PvuII and dephosphorylated with shrimp alkaline phosphatase to avoid self-ligation. Then the amplified and filled fragment (Prrn+RBS+aadA+rbcL) was blunt-end cloned in the middle of the 3 kb sequence (between INSL and INSR) into the pUCINS to yield the pCSI vector.
3.6. Transformation of E. coli Using Heat-Shock Method
For transformation of recombinant vector to E. coli, 4 µl of ligation reaction mixture was added to the competent cells of E. coli strain DH5α, gently stirred to get a uniform mixture and then was kept on ice for 30 minutes. A heat shock was applied in a 42°C water-bath for 90 seconds, and then was put on ice immediately for 2 minutes. 900 µl of SOC medium was added and incubated on a shaker incubator in 200 rpm, at 37°C for an hour. A proper volume of the culture was spread on LB plates with appropriate concentrations of antibiotics (75µg/mL ampicillin). The plates were inverted after the liquid has been absorbed, and were placed in a 37°C incubator overnight.
4. Results
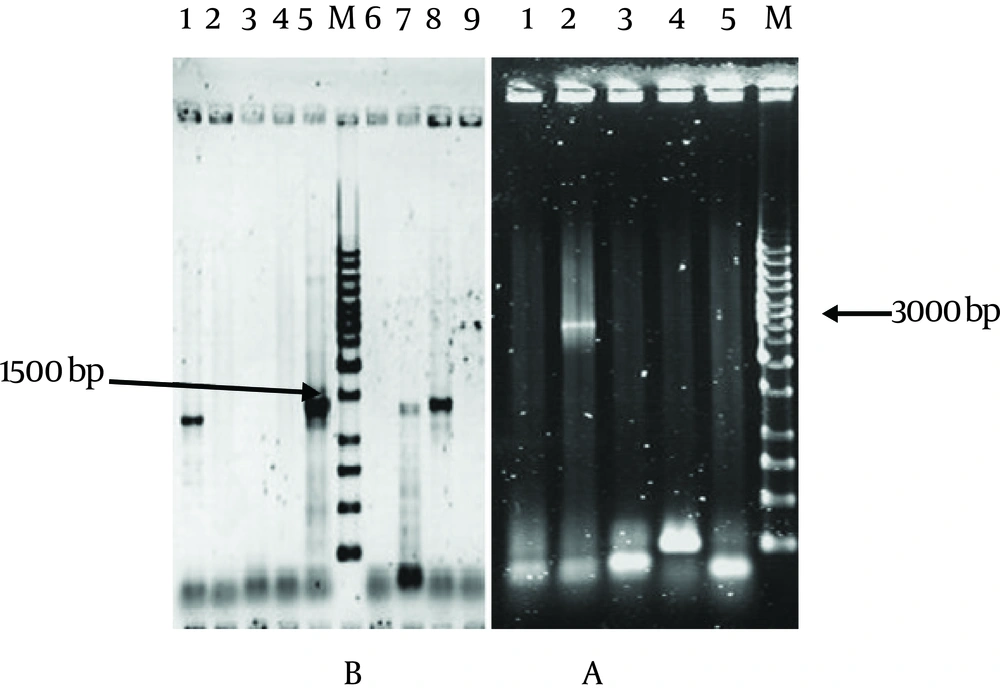
An appropriate integration site was allotted in the present study. The spinach plastome was completely sequenced and available in public databases. The region between the trnI and the trnA genes was selected from spinach plastome, and then specific primers were designed and synthesized. A 3000-bp-long fragment was amplified via PCR and then was blunt-end ligated into the PvuII site of vector pUC19. The pUCINS construct was transformed to E. coli using heat-shock method and the resulting clones were confirmed via several methods including colony PCR ( Figure 1A ), digestion and sequencing.
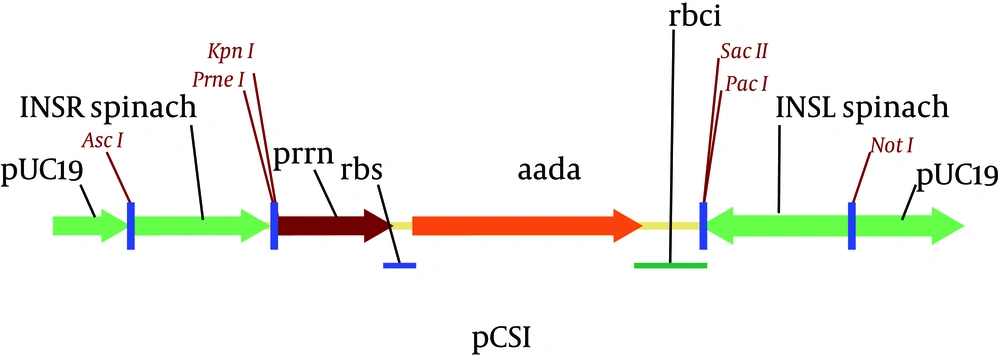
The selectable gene, aadA, under the control of tobacco and Chlamydomonas plastid regulatory elements was fused to a short synthetic ribosome-binding site (RBS). This fragment was also amplified via PCR using specific primers and then was blunt-end ligated into the pUCINS between the cloned INSL and INSR sequences via PvuII restriction site. The constructed plasmid map of pCSI has been shown in Figure 2 .
A. Proof of the presence of INSL and INSR (3 kb sequence) in pUCINS, M: 1 kb size marker, Lane: Negative control (pUC19), Lane 2, 3, 4, 5 : Random selected colonies. Colony 2 shows a band in 3000 bp. B. Proof of the presence of aadA gene by colony PCR technique. M: 1 kb size marker, Lane1, 2, 3, 4, 5, 7, 8 and 9: Random selected colonies. Lane 6: Negative control (pUCINS without gene).
The capacity of the aadA gene to confer resistance to the streptomycin in E. coli was investigated. pCSI vector consists of the aadA gene which was under the control of Prrn promoter and rbcL terminator. The bacterial culture of this construct was spread on LB plates containing 75µg/mL ampicillin and various concentrations of streptomycin, then incubated at 37°C overnight. The resistance to streptomycin was estimated by the appearance of bacterial cell growth on the plates as listed in Table 1 .
| Trial No. | Concentration, µg/mL | DH5α (E. coli) with pUC19 | DH5α (E.coli) Without Plasmid | DH5α (E.coli) With pCSI | |
|---|---|---|---|---|---|
| Ampicillin | Streptomycin | ||||
| 1 | 75 | 80 | - | - | ++++ |
| 2 | 75 | 100 | - | - | ++++ |
| 3 | 75 | 160 | - | - | ++ |
| 4 | 75 | 240 | - | - | - |
| 5 | 75 | 320 | - | - | - |
5. Discussion
In the present study, an investigational report of bacterial aadA gene which is controlled via the tobacco chloroplast Prrn promoter and the Chlamydomonas rbcL 3΄UTR has shown a successful expression in E. coli.
As mentioned above, the transgene integration in the chloroplast genome performs via double homologous recombination (1, 19). Such mode can help plastid vectors to carry left and right flanking sequences each with 1 to 2 kb in size from the host plastid genome (8). The insertion sites of foreign genes have been well documented in several crops, including rbcL-accD, trnV-rps12/7, trnI–trnA, rps7-ndhB and ndhF-trnL intergenic regions (10). To date, one of the most commonly used site of integration reported is region between the trnI and the trnA genes, within the rrn operon, located in the IR regions of the chloroplast genome which is able to induce two copies of transgene per plastome (9).
The foreign gene expression levels obtained from gene integration at this site were highest ever been reported (6). So this site was selected for the spinach chloroplast vector as flanking regions. Both left and right flanking sequences that were chosen from spinach plastome, were 1500 base pairs. These sequences were amplified and then cloned to the pUC19. This length is enough for proper homologous recombination. Using longer sequences leads to larger plasmids which may decrease the efficiency of chloroplast transformation. On the other hand, having plastome with shorter sequences could decrease the homology and consequently reduce the rate of heterologous recombination. Selectable marker genes are essentially important to provide uniform transformation for all genome copies. In this report, the aadA gene was employed as a selectable marker gene which can contribute to spectinomycin and streptomycin resistance in prokaryotic and eukaryotic cells and frequently can be used in chloroplast transformation. The aadA gene amplified via PCR and cloned in pUCINS under the control of Prrn promoter and rbcL terminator.
In this investigation, the aim was to examine the efficacy of this vector in E. coli strain DH5α. When bacteria added on media containing 80, 100 and 160 mg/liter streptomycin, the bacteria could grow, while E. coli strain DH5α containing pUC19 did not show any growth on those media. These findings indicated the bacterial cells capability to express the aadA gene under the control of tobacco and Chlamydomonas plastid regulatory elements and induce relatively high resistance to streptomycin. These results approve that the transcription and translation systems of bacteria can recognize the chloroplastic elements. In conclusion, the findings indicated that the chloroplast tobacco Prrn promoter and Chlamydomonas rbcL terminator were capable of transcription and translation of the aadA gene in bacteria. Such mechanism is believed to be relevant to endosymbiosis contemplate evolutionary.
The data presented in this paper contains the first investigational report regarding the construction of chloroplast vector for spinach plant. In future, this system can be used as a valuable tool for agronomical and biotechnological relevant traits induction into spinach plant.