1. Background
Influenza A and B viruses are the major causative agents of acute respiratory infection, responsible for a high level of morbidity and mortality, particularly among infants, elderly, individual with pulmonary and cardiac diseases, diabetes, and patients with immunocompromised status worldwide (1, 2). The World Health Organization (WHO) estimated that 250000 to 500000 deaths occur due to influenza A and B viruses annually (3). Influenza A and B viruses belong to the Orthomyxovirus family which possesses eight segmented single-stranded RNA (4).
The influenza viruses types can be distinguished on the basis of antigenic differences in nucleoprotein and matrix proteins (5). In addition, influenza A viruses are divided into several subtypes based on the antigenic differences on the external glycoprotein, the hemagglutinin (HA), and the neuraminidase (NA) proteins (6). Until now 16 HA [H1–H16], and 9 NA [N1–N9] subtypes have been detected (7). HA and NA proteins of influenza type B are more stable than the type A, HA, and NA (5). Co-infection of influenza A/H1N1 and A/H3N2 with influenza B have been demonstrated in human population (6).
Routine lab tests for the diagnosis of influenza are serology and antigen detection (1). The common methods for subtyping influenza viruses include growth of virus in cell culture (i.e. MDCK) or embryonated chicken eggs prior to typing and subtyping by hemagglutination inhibition (HI), Enzyme-Linked Immunosorbent Assay (ELISA), or immunofluorescence (IF) (5). The main limitations of these techniques are time-consuming, low sensitivity, and low specificity. The use of nucleic acid amplification techniques has been found to be more sensitive for determining the type and subtype than the standard tissue culture (1).
2. Objectives
To date there is no report of epidemiological description of influenza viruses in Khuzestan province. Therefore for the present study the reverse transcription RT-PCR assay was used for typing and subtyping influenza viruses in clinical samples during 2009-2010. Khuzestan province with the capital city of Ahwaz is located in the southwest of Iran. The population of Khuzestan province is about 4.5 million.
3. Patients and Methods
3.1. Virus Strains and Clinical Specimens
The study was approved by the Ethical Committee of Ahvaz Jundishapur University of Medical Sciences. The decision to admit patients for this study was performed according to the clinical condition of patients. Patients who fulfilled the case definition criteria in province of Khuzestan, Iran during March 2009 to February 2010 were included in this study.
Stock strains of influenza A and influenza B virus were obtained from the virology department of Public Health School, Tehran University Medical Science. The viruses used were the A/New Caledonia/20/99 H1N1, A/Panama/2007/99 H3N2, and B/Shangdong/7/97.
Nasopharyngeal swab specimens were obtained by physicians by a Dacron swab with a plastic shaft from 655 patients with influenza-like symptoms including fever, coughing, rhinorrhea, myalgia, arthralgia, and diarrhea. Patients’ age was between 1 month and 85 years with the average age of 25.07±18.24. The most suspected patients with flu-like symptoms were observed from Ahvaz (49.5%), and the lowest cases were seen in Baghmalek (0.01%). Clinical samples were taken within the 48 hours of initial symptoms. All the clinical samples were collected in the transport median (Copan Italia, Brescia, Italy) containing Hank's Balanced Salts, Bovine Serum, Albumin L-Cysteine, Gelatin Sucrose, L-Glutamic Acid, HEPES Buffer, Vancomycin, Amphotericin B, Celestin, Phenol Red. All the samples were stored at -70 ͦ C prior to the test.
3.2. Nucleic Acid Extraction and cDNA Synthesis
Viral RNA was extracted from 200 μl of the each clinical sample using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instruction. The RNA was immediately kept at -70 ͦ C. Complementary DNAs synthesis was performed using M-MLV reverse transcriptase (Fermentas, Burlington, Canada), and random hexamer primers (Fermentas, Burlington, Canada) according to the manufacturer’s instructions.
3.3. Polymerase Chain Reaction
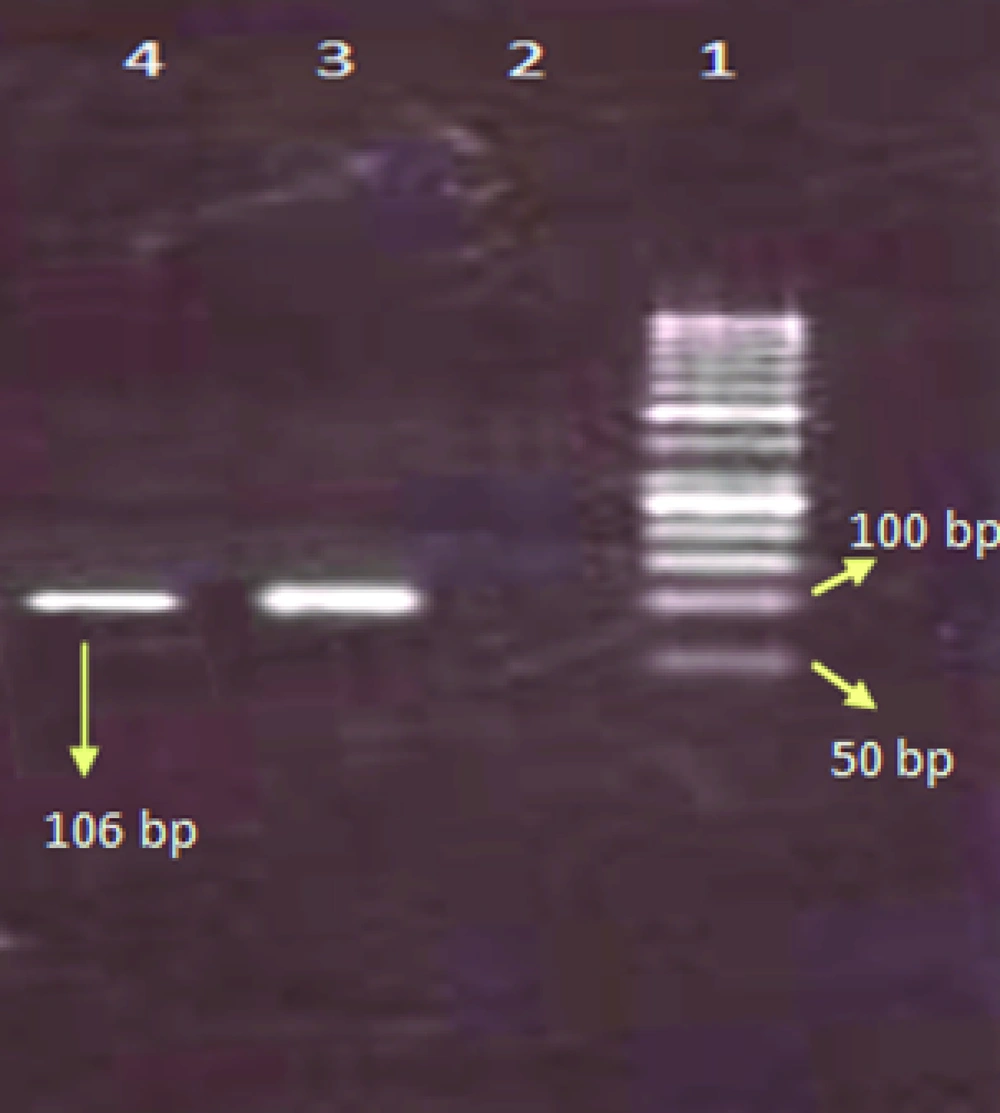
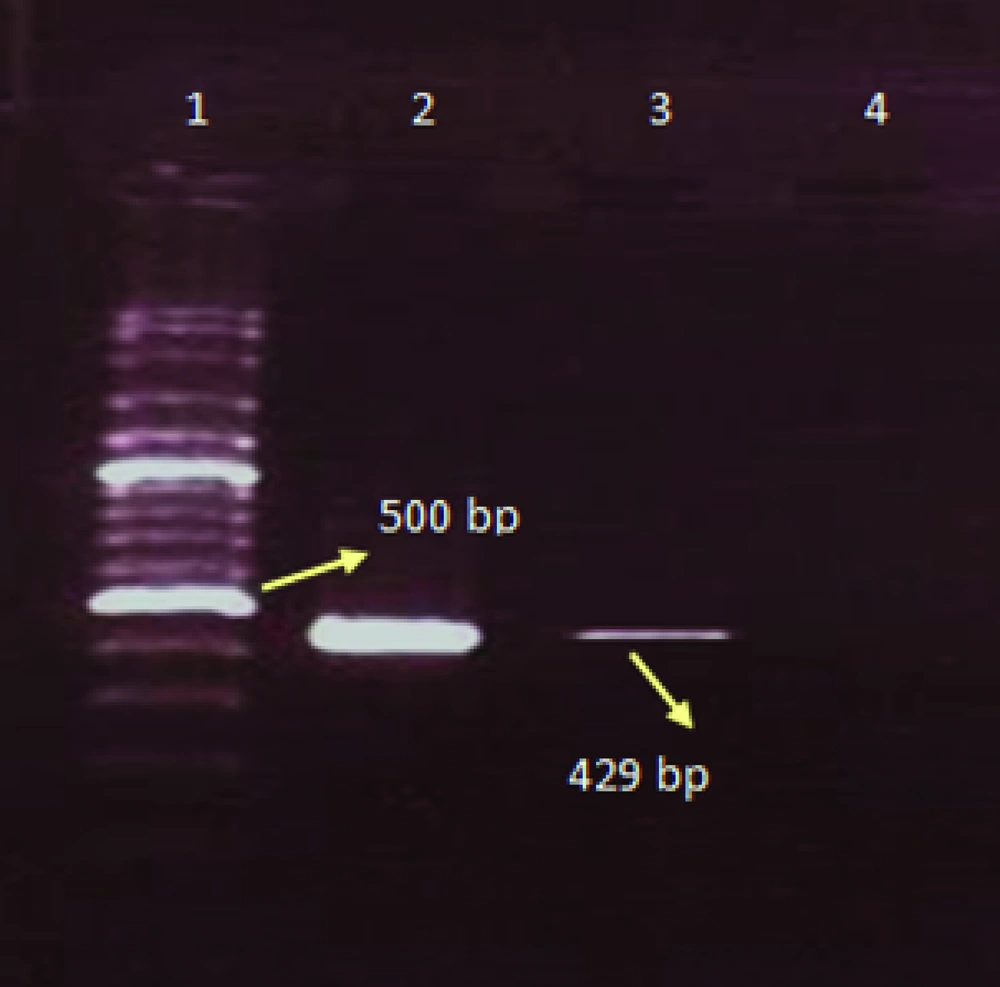
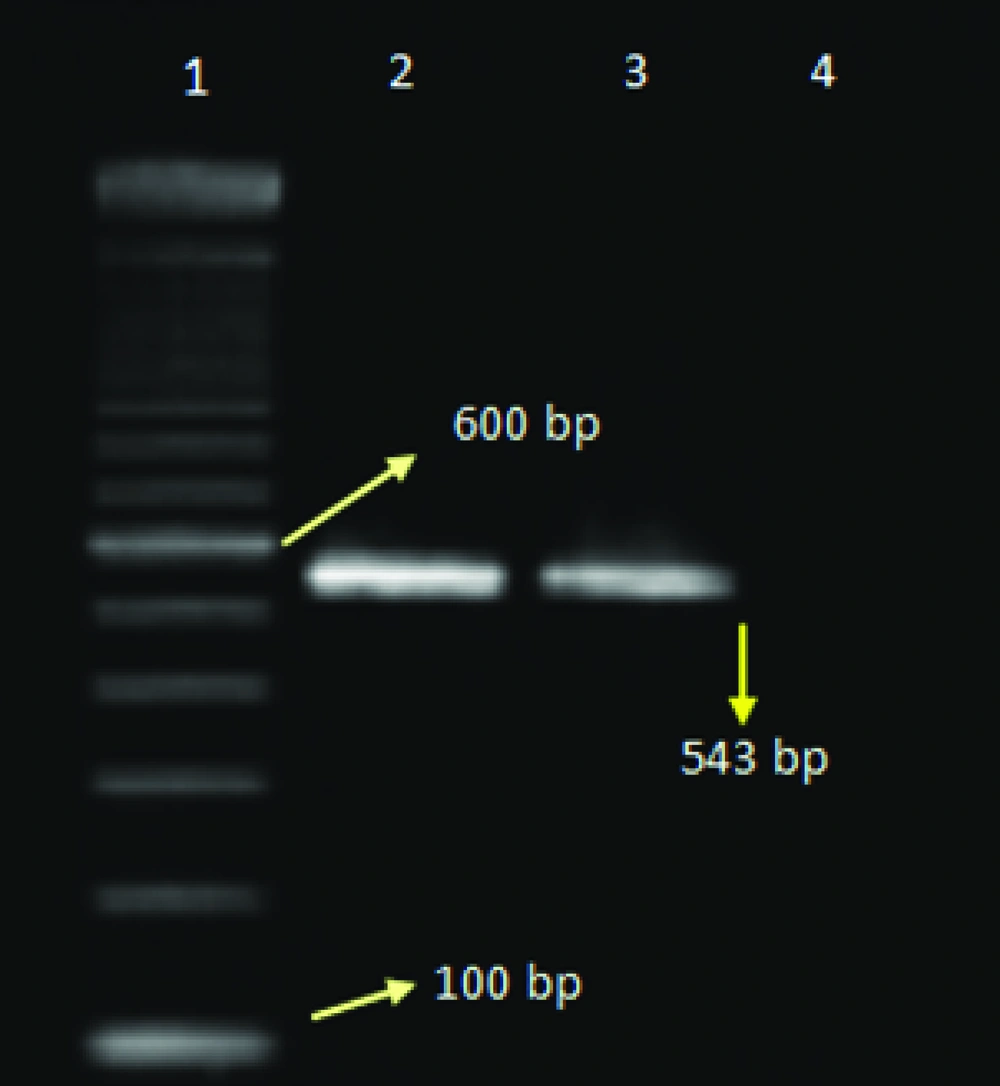
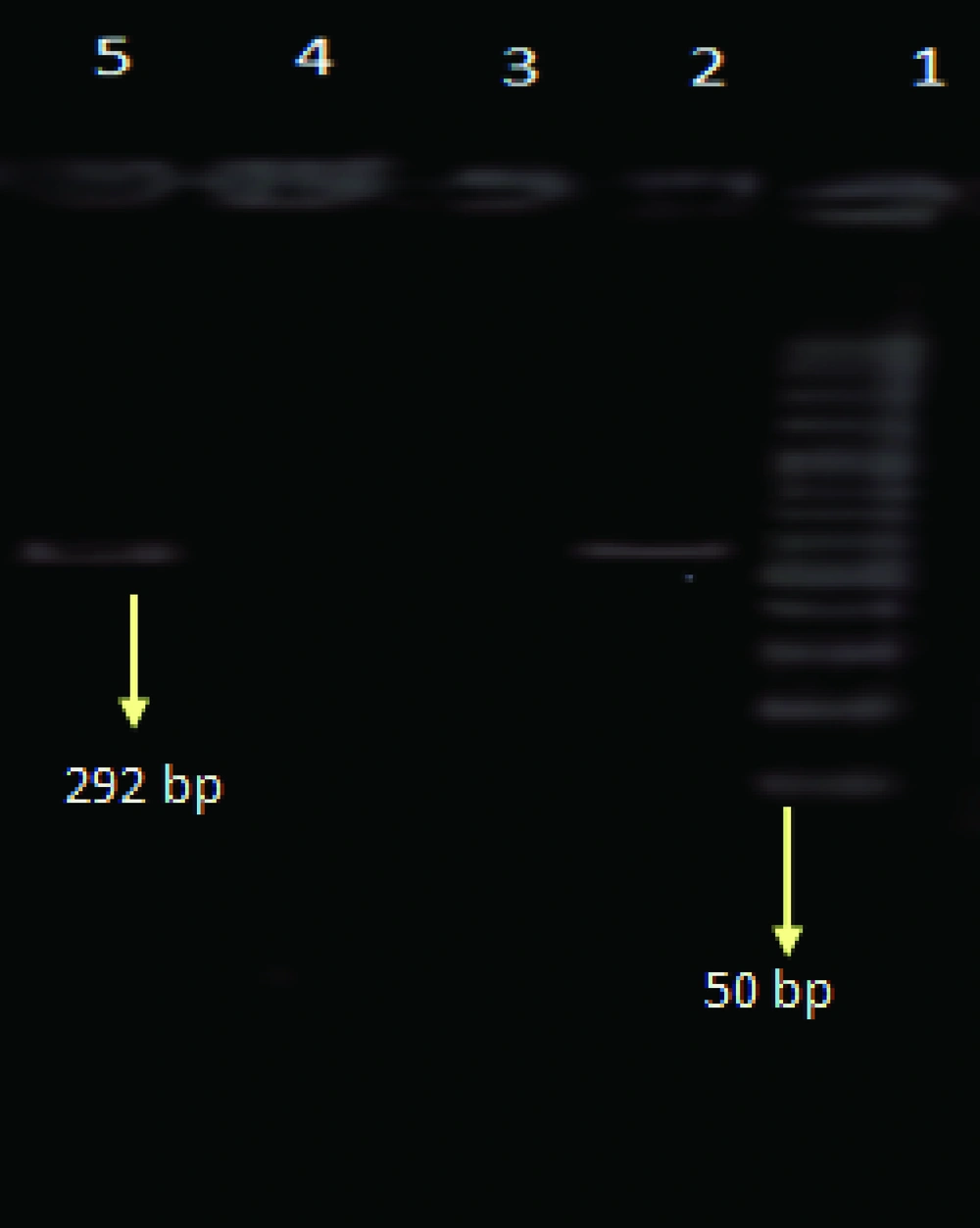
PCR was performed for four different reactions separately including influenza A, H1N1, H3N2, and B viruses. Initially PCR test was performed for the determination of influenza A and B viruses, later the samples with positive results for influenza A virus were tested for the detection of influenza A/H3N2, and A/H1N1. For the determination of influenza A virus the matrix primers, and for influenza B virus the nucleoprotein primers were used ( Table 1 ). For influenza A subtypes the hemagglutinin gene was used ( Table 1 ).
| Name | Sequence (5′→3′) | Gene Target-Nucleotide Position | Amplicon (bp) | Reference |
|---|---|---|---|---|
| Flu-M-F | GACCRATCCTGTCACCTCTGAC | 91→112 | (8) | |
| Flu-M-R | AGGGCATTYTGGACAAAKCGTCTA | 196→173 | 106 | (8) |
| NP-B-For | ATAGGAAGAGCAATGGCAGACAGAG | 72–796 | (8) | |
| H1-A-For | CCGACTATGAGGAACTGAGGGAG | 335–375 | (9) | |
| H1-A-Rev | TCGCATCACATTCATCCATTG | 857–877 | 543 | (9) |
| H3-A-For | TGAACGTGACTATGCCAAACAATG | 547–570 | (9) | |
| H3-A-Rev | CGTATTTTGAAGTAACCCCGAGGA | 815–838 | 292 | (9) |
6 μl of cDNA was added to 19 μl of master mix containing 1x PCR buffer, 1.5mM MgCl2, 0.2mM dNTPs (Fermentas, Burlington, Canada), 1.5 U Taq polymerase enzyme (CinnaGen, Tehran, Iran), and 0.25mM of each primers (Bioneer, Seoul, South Korea). Amplification was performed for 35 cycles. The PCR reaction mixture was subjected to thermal cycler (Techne, Cambridge, UK) within the initial program at 94°C for 5 minutes, as follows: denaturation for 45 sec at 94°C, hybridization at 51°C (for MA), 54°C (for NP-B primers), 52.5°C (for H1), and 54.5°C (for H3 primers) for 45 sec, elongation at 72°C for 45 sec, and a final extension at 72°C for 7 min (9). PCR products were visualized by ethidium bromide following electrophoresis on 2% agarose gel.
3.4. Statistical Test
Statistical test was performed using SPSS version 17.0 (SPSS Inc, Chicago, Illinois).
Comparisons were performed between male and female using Х2 test in patients with positive and negative results for influenza. P < 0.05 was considered statistically significant.
4. Results
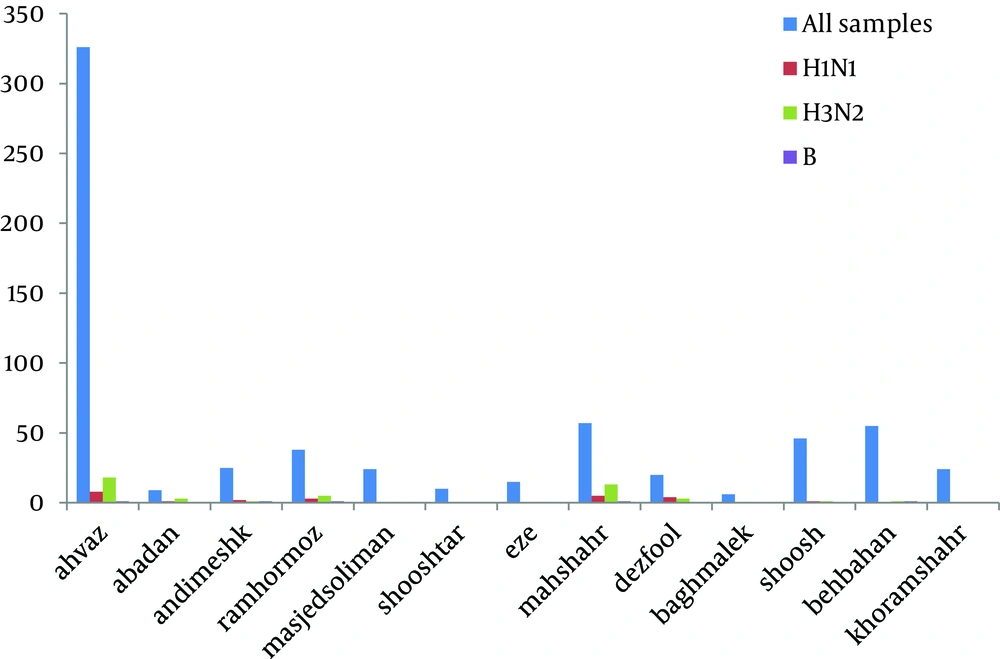
The total prevalence of seasonal influenza A and B viruses was 11.29%. Of 655, 69 (10.53%) samples had positive results for human influenza A virus, and 5 (0.7%) samples had positive results for influenza B. Of those 69 samples with positive results for influenza A virus, 45 (6.8%) belonged to H3N2, and 24 (3.6%) to H1N1 subtypes. Agarose gel electrophoresis of PCR products are shown in Figures 1,2 ,3, and 4. The sample positive included 49% males and 51% females ( Table 2 ). The highest prevalence of seasonal influenza observed in Ahvaz city 4% (27), and Mahshahr city 2.5% (19). Although, influenza A and B were not detected in Masjedsoleiman, Shooshtar, Izeh, Baghmalek, and Khoramshar cities (Figure 5).
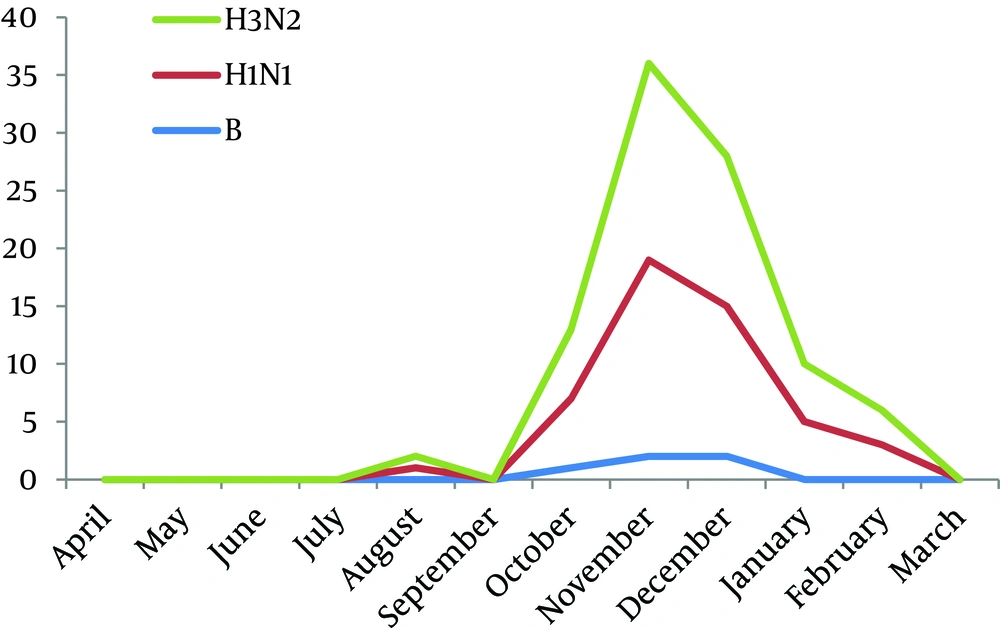
The average age of patients with positive results for influenza viruses was 20.18 ± 16.22 years old, compared to 25.87 ± 18.80 years old for those with negative results for influenza viruses. Most positive cases were found in patients aged less than 20 years. The duration of seasonal influenza was 7 months (Figure 2). The peak of seasonal influenza was between October and December 2009. The first cases were observed during mid August (Figure 6).
| Spimens, NO. | Type | Subtype A | |||
|---|---|---|---|---|---|
| Age, y | Infuenza A | Ifluenza B | H1N1 | H3N2 | |
| 0-10 | 138 | 22 | 1 | 4 | 18 |
| 11-20 | 148 | 26 | 0 | 13 | 13 |
| 21-30 | 160 | 12 | 1 | 3 | 9 |
| 31-40 | 85 | 4 | 0 | 2 | 2 |
| 41-50 | 49 | 4 | 1 | 1 | 3 |
| 51-60 | 41 | 1 | 1 | 1 | 0 |
| >60 | 31 | 0 | 1 | 0 | 0 |
| Total | 652 | 69 | 5 | 24 | 45 |
| Gender | |||||
| Male | 354 | 33 | 2 | 12 | 21 |
| Female | 301 | 36 | 3 | 12 | 24 |
| Total | 655 | 69 | 5 | 24 | 45 |
5. Discussion
Confirming the diagnosis of influenza with rapid tests such as RT-PCR is important for the clinical management to reduce the disease severity, and the early use of antiviral drugs (10). Furthermore, rapid diagnosis and accurate identification of current strains of influenza virus have epidemiological value. Another value of a rapid laboratory diagnosis that cannot be underestimated is for subsequent annual vaccine. Every year the WHO recommends vaccine strains for the forthcoming seasonal influenza, and for preventing epidemics based on the surveillance data gathered worldwide (11).
In this study the prevalence of type and subtypes of human influenza A were assessed in Khuzestan province of Iran during 2009–2010. There are several reports on the prevalence of influenza in Iran. In Tehran, Soltani and colleagues reported in 2007 that 12 of 57 samples (21%) had positive results for human influenza using RT PCR, which 7 were A/H3N2, 3 were A/H1N1, and 2 were B Subtypes (10). Mousavi et al. in 2008-09 in Tehran reported that of 142 samples, 17 had positive results for seasonal influenza virus, which 15 were H1N1, and only 2 H3N2 were detected, and influenza type B was not identified (12).
In Malaysia, Zainah Saat and colleagues in 2009 reported13.5% (227/1,678) seasonal Influenza, which 78.0% (177/227) influenza A, and 22.0% (50/227) influenza B were typed and subtyped by hemagglutination inhibition (HAI) assays (11). In Western Australia Dale Carcione et al. reported 11.2% seasonal influenza by RT PCR in 2009 (13). Ji Rong Yang and colleagues in 2009 in Taiwan reported 14.11% seasonal influenza which (13.4%) H3N2, 12 (0.7%) H1N1 and 1 (0.06%) influenza B viruses were identified by real-time RT-PCR (2). Our findings showed that the rate of positive influenza virus was the same as the range described in Malaysia, Taiwan, and Western Australia studies (2, 11, 13).
The prevalence of positive seasonal influenza among male (49%) and female (51%) was not found significant (P > 0.05). Dingle et al. reported that the highest attack rate of seasonal influenza was observed in school-age children, and they might have a critical role in disseminating seasonal influenza (14). However in the present study the highest prevalence of seasonal influenza was observed in two age groups: one group with 31% (23/138) less than 10 years old, and the second group with 37% (28/148) aged 11-20 years old. The lowest prevalence of seasonal influenza was observed among patients over 60 (1/31) (3%).This finding revealed the increased distribution of the seasonal influenza viruses in young age groups during 2009-2010.
Seasonal influenza occurs in winter in the Northern hemisphere, but its peak of activity time is varying. The "peak month of seasonal influenza activity" occurs in January or later (15). The Center for Disease Control (CDC) reported that, during 2009-2010 seasonal influenza began earlier causing pandemic H1N1, and activity peaked in October and next declined in January. In agreement with this finding in the present study the month of influenza peak activity was observed during October and December 2009, which seems to be correlated with the US CDC surveillance data (15).
This study has some limitations, including lacks of patients' demography such as history of allergic disease, pregnant woman, patients with renal failure or cardiac problem, and individual with immune deficiency which requires further investigation. The suspected influenza patients admitted in private clinics were not included in the present study. In this study 88.71% of samples, had negative results for influenza viruses. Besides, other viruses including Metapneumovirus (16), Adenovirus, Respiratory Syncytial virus, and Parainfluenza (17, 18) viruses which can cause the influenza-like symptoms were not studied in the present study. Finally, regarding to prevalence of influenza in Khuzestan, the immunization program could significantly decrease the number of the influenza infection, particularly among children and elderly.